Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Genome Changes Mapped In Humans
Epigenomics: Most comprehensive results to date in collaborative epigenome-mapping project
by Stu Borman
February 19, 2015
| A version of this story appeared in
Volume 93, Issue 8
Researchers reported last week that a DNA-methylation gene called uhrf1 is a molecular trigger for inflammatory bowel disease (Proc. Natl. Acad. Sci. USA 2015, DOI: 10.1073/pnas.1424089112). It was already known that immunity and genetics play a role in the disease. “But we’ve found that it is not just the immune genes themselves but also the regulation of those genes through epigenetics that can cause problems,” says corresponding author and Duke University cell biologist Michel Bagnat.
The finding is just one example of the importance of epigenetics, or epigenomics—the study of how modifications to a cell’s genome can influence human development, health, and disease. If a series of studies published last week in Nature and several other Nature research journals are any indication, there will be many more such examples to come (http://nature.com/epigenomeroadmap).
The studies report the most comprehensive results to date from the Roadmap Epigenomics Project, the epigenomics version of the Human Genome Project.
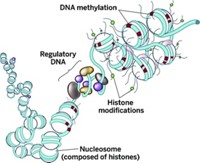
All the nongerm cells in a particular organism are genetically identical, but not all genes get expressed the same way across cell types. Such variations in gene expression are what differentiate embryonic from adult cells, heart from brain cells, and normal from diseased cells. The epigenomic factors that control differential gene expression include methylation of specific DNA bases, chemical modifications to the proteins called histones that package eukaryotic DNA, and DNA conformation changes that promote the binding of regulatory transcription factors.
Researchers in the Roadmap Epigenomics Project—an international collaboration sponsored by the National Institutes of Health—have now mapped the epigenomes of more than 100 human cell types and report on how epigenomics affects diseases such as cancer and Alzheimer’s, differentiation of stem cells into adult cells, and other biological functions.
In a Nature commentary, molecular biologist Hendrik G. Stunnenberg of Radboud University, in the Netherlands, says that the community now “plans to determine the epigenomes of every cell type in the human body—estimated to be several hundred to 1,000.”





Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter