Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Chemical reaction lights the way for tracking microRNA in living organisms
Imaging probes provide first-ever technique for visualizing small RNA sequences that control gene expression in vivo
by Alla Katsnelson, special to C&EN
June 13, 2016
| A version of this story appeared in
Volume 94, Issue 24
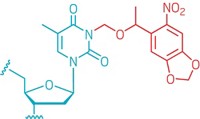
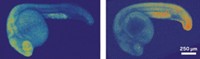
Researchers have developed a light-induced chemical reaction that visualizes RNA in live zebrafish embryos without interfering with cell processes. In previous work, the team applied the method to cells in culture. But now the researchers have developed the method as the first technique for detecting specific strings of nucleic acids in live vertebrates that doesn’t require genetically modifying the organisms. What’s more, the method is sensitive enough to visualize the expression of microRNAs, which are small noncoding RNAs that act as puppet masters of gene expression (ACS Cent. Sci. 2016, DOI: 10.1021/acscentsci.6b00054). Nicolas Winssinger, Marcos Gonzalez-Gaitan, and colleagues at the University of Geneva designed two nucleic acid probes that each complement and bind to adjacent halves of a target microRNA sequence. The researchers conjugated one probe to a ruthenium complex that absorbs visible light and the other to a rhodamine dye that lights up when its azide bonds are cleaved. When the probes attach to the target sequence, the two reagents come close enough to react. Shining a light on the sample activates the ruthenium, which reduces the azide in the rhodamine conjugate and thereby allows it to fluoresce. The researchers designed probes against three different microRNAs: one expressed in neural cells, another in muscle cells, and a third in cells involved in forming the pectoral tail and fins. After the embryos were exposed to light for 30 minutes, confocal imaging revealed that each probe set illuminated the expected type of tissue and the probes tracked microRNA expression over time during embryo development.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter