Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Scientists settle debate over how a potassium ion channel works
Potassium ions alternate with water molecules as they pass through channel, study shows
by Jyllian Kemsley
September 26, 2016
| A version of this story appeared in
Volume 94, Issue 38

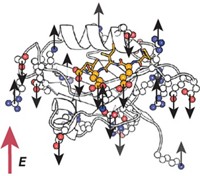
Ubiquitous in living organisms, potassium ion channels help provide energy to cells. In excitable cells such as neuron and muscle cells, the proteins also function to transmit signals from one part of a cell to another.
Despite more than 50 years of research, however, questions still remain about exactly how these channels do their job. Scientists do know that the channels, which sit in cell membranes, establish an electric potential between the inside and outside of cells by tightly controlling how many potassium ions pass through the protein. Precisely what mechanism they use to transport those ions has been up in the air.
A new interdisciplinary approach is starting to find answers, recently demonstrating that such transport involves alternating potassium ions and water molecules (Science 2016, DOI: 10.1126/science.aag1447).
The approach involves partially synthesizing a potassium ion channel to incorporate site-specific isotopic labels. Researchers then study the labeled protein in solution using two-dimensional infrared spectroscopy and compare the spectra they collect with those simulated from molecular dynamics computations.
“It’s a fantastic example of something that simply could not be done by one lab,” comments Merritt Maduke, a professor of molecular and cellular physiology at Stanford University who studies ion transport proteins. “There are a lot of membrane-protein structures out there. But the reality is that in order to really understand how the channels work, we need to see them in motion,” Maduke adds.
In potassium channels such as KcsA—the one studied in the new work—K+ ions transit through a narrow pore known as the selectivity filter. The filter is lined by carbonyl groups that serve to make four K+ binding sites. The mechanism that scientists have long believed to be at work in KcsA and other channels is known as the “knock on” model: At any one time, two K+ and two H2O molecules are bound alternately in a single-file column in the filter. The approach of another K+ from the inside of the cell causes the column to advance, pushing a K+ and an H2O out of the cell as another pair enters.
But in 2014, a molecular dynamics study led by Bert L. de Groot of the Max Planck Institute for Biophysical Chemistry, Ulrich Zachariae of the University of Dundee, and Chen Song of Oxford University suggested an alternative, “hard knock” mechanism (Science 2014, DOI: 10.1126/science.1254840). The hard-knock model has only K+ ions going through the channel. An entering K+ collides directly with ions already in the channel, pushing one toward the exit. Simulations of both the knock-on and hard-knock models reproduced existing experimental data equally well.
To determine which mechanism is accurate, a new team formed, led by protein semisynthesis expert Francis I. Valiyaveetil of Oregon Health & Science University; spectroscopists Martin T. Zanni of the University of Wisconsin, Madison, and Eduardo Perozo of the University of Chicago; and computational chemist Benoît Roux of the University of Chicago.
To create isotopically labeled KcsA, Valiyaveetil and coworkers expressed in Escherichia coli two recombinant peptides about 70 amino acids long, one each from the N- and C-terminal ends of the KcsA channel. Separately, they synthesized a 12-amino acid piece that would go in between, with 13C and 18O labels added to three protein backbone carbonyls that help make up the selectivity filter. To complete the semisynthesis, they ligated the three pieces together and folded the protein. “The overall procedure was pretty straightforward,” Valiyaveetil says, although his group had to scale up and optimize procedures to ensure good yields.
The protein samples went from Valiyaveetil’s lab to Zanni’s for 2-D IR spectroscopic studies.
One advantage of 2-D IR over other methods is that the IR technique boasts a time resolution of one to two picoseconds, which is faster than proteins transition between conformations. “Nothing moves except the water, and so the 2-D IR spectra give an instantaneous snapshot of the structures in your solution,” Zanni says. That allows comparison to spectra calculated from molecular dynamics simulations to test proposed structures or dynamic trajectories.
Another advantage is that atomic bond vibrations measured by 2-D IR are sensitive not just to protein structures but also nearby ions and water molecules. The isotopic labels on KcsA caused a shift in the frequency of the protein’s carbonyls. The carbonyl frequencies also shifted when ions were in the channel, allowing 2-D IR to probe the transport mechanism in real time.
“We had to be very careful to subtract off the background of the natural protein,” but otherwise the data collection followed standard procedures, says lead author Huong T. Kratochvil, a graduate student in Zanni’s lab.
The result was an unprecedented exploration of the ion channel in action. Comparing the experimental spectra to those computed for the two possible mechanisms, the researchers conclusively determined that the channel follows the knock-on model of alternating K+ and H2O transport. They also found that a valine carbonyl flips in and out of the pore, something that had been suggested by previous molecular dynamics simulations but not observed in crystal structures.
“This is a really fantastic example of what 2-D IR spectroscopy can tell you about the function of even gigantic membrane proteins,” comments spectroscopist Andrei Tokmakoff, a chemistry professor at the University of Chicago. “This is what I call a ‘blue sky’ experiment—it’s the type of study that scientists always hope to accomplish after years of effort.”
“I’m excited that by using 2-D IR to test the simulations, we could bring a new approach to the ion channel field and solve an outstanding dilemma,” Zanni says.
And because semisynthesis is applicable to a range of membrane proteins, “there are many more questions in the ion channel community that I want to address with this technique,” Valiyaveetil adds.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter