Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Multifaceted molecule casts a universal glow on cell surfaces
A new fluorescent probe binds to and illuminates the surfaces of bacterial, fungal, and mammalian cells
by Erika Gebel Berg
May 20, 2016

Commercial probes that light up cell surfaces can be expensive and picky, preferring certain cell types over others. Now, researchers have developed a simple imaging molecule that readily binds to mammalian, fungal, and bacterial cells, thanks to the right combination of properties (ACS Biomater. Sci. Eng. 2016, DOI: 10.1021/acsbiomaterials.6b00130).
Fluorescent molecules that bind to a cell’s surface allow researchers to observe its structure. This can reveal whether a cell is healthy, dying, or dividing; how drugs interact at the surface; or how drug-delivering particles transfer their payload. Commercial cell-surface dyes are available, but cells quickly engulf these probes, says Zhan Chen of the University of Michigan, Ann Arbor, limiting the amount of time the cells can be imaged.
In a previous study, Chen’s team designed a longer-lasting molecule that binds to the cell surface at multiple points, making it harder for the cell to consume it. The researchers soon realized that the new molecule had a bonus feature: It bound not only to the plasma membranes of mammalian cells, but also to the cell walls of yeast and bacteria. In the new study, Chen, along with Fu-Gen Wu of Southeast University, in China, and colleagues, uncovered how the molecule achieves its universal cell appeal.
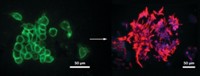
The cell imaging probe is “like a long rope with smaller ropes attached along its length,” Chen says. The main rope is glycol chitosan, a positively charged polysaccharide. The smaller ropes are polyethylene glycol (PEG), each capped at the end with a molecule of cholesterol. The glycol chitosan backbone is also dotted with a dye, fluorescein isothiocyanate, which lights up the cell surface for viewing with confocal microscopy.
To figure out how the probe’s components enable it to bind different cell types, the researchers synthesized test molecules, one missing the probe’s PEG-plus-cholesterol ropes and another without the glycol chitosan. The researchers treated bacterial, fungal, or mammalian cells with each of the test molecules and looked to see if the cells still glowed. The test probe missing the hydrophobic cholesterol molecules failed to illuminate the mammalian cells, while the one without a positively charged backbone didn’t bind the bacterial or yeast cells. Chen says this outcome stems from the differences between the surface properties of cell membranes versus cell walls. The hydrophobic interaction with cholesterol is mainly responsible for the probe’s binding to the mammalian cells, he says. Yeast and bacterial cell walls are highly negatively charged, so it follows that the probe’s positively charged backbone anchors it to these cells.
“Current labeling techniques can in some cases be complicated, pricey, and can also be specific to the cell type of interest,” says Virginia C. Spanoudaki, a cell-imaging scientist at Massachusetts Institute of Technology. “Having a fast and cheap labeling technique to understand cell interactions is valuable.” The researchers have yet to demonstrate that the method could be used for high-throughput cell imaging, she adds, which would make the probe especially useful.
Chen plans next to examine whether the probe can be modified to target drugs to cell surfaces.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter