Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Computationally designed ‘mini proteins’ prevent flu infection in mice
High-throughput design technique could lead to novel therapeutics
by Stu Borman
September 28, 2017
| A version of this story appeared in
Volume 95, Issue 39

Scientists have used a computational method to design brand-new small proteins that inhibit specific therapeutic targets such as the hemagglutinin protein involved in flu infection. The findings suggest that this method—de novo protein design—could be a wave of the future for drug discovery.
Researchers often find drugs by screening large collections of compounds for those that happen to hit biological targets. Using computers to intentionally design compounds to hit targets is a newer and less proven strategy. A key advantage of this route to drug discovery is that its underlying prediction algorithms can be improved over time, whereas screening is essentially random.
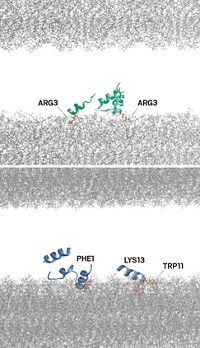
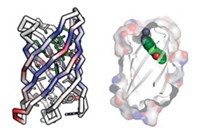
David Baker of the University of Washington and coworkers demonstrated the potential of this new approach by designing therapeutic “mini proteins,” each containing about 40 amino acids (Nature 2017, DOI: 10.1038/nature23912). In cell culture experiments, the proteins inhibited hemagglutinin and botulinum neurotoxin B, the cause of botulism. And one mini protein, called HB1.6928.2.3, prevented flu infection in mice when administered by inhalation within 72 hours of exposure to the virus.
Baker and coworkers, including Aaron Chevalier, Daniel-Adriano Silva, and Gabriel J. Rocklin, used the group’s computer software, Rosetta, to design nearly 23,000 mini proteins, about 100 times as many as in previous de novo protein design studies. Rosetta assembled the mini proteins from component parts—α-helices, β-sheets, loops, and disulfides—by predicting folded shapes likely to interact with known target sites on hemagglutinin or botulinum toxin.
The researchers used Rosetta to evaluate the resulting mini proteins’ binding properties. Then they synthesized DNAs that code for about 10,000 of the best designs and inserted the DNAs into yeast, which expressed the corresponding proteins on their surfaces. The scientists screened the yeast experimentally to see which of the proteins displayed on their surfaces bound most tightly to the flu and botulinum targets. The researchers then sequenced those proteins and fed information about them back into Rosetta to improve the software’s predictive ability.
Two iterations of this design process yielded 14 flu and botulinum inhibitors. These mini proteins are easier to design and synthesize, more heat stable, and potentially less immunogenic than antiflu antibodies, such as those used in vaccines. But the mini proteins aren’t orally available like many small-molecule drugs and are a long way from being clinical candidates.
Many drug discovery efforts have used high-throughput screening, “while the advantages of computational methods have largely been overlooked,” but the new study and others suggest that computer design is coming to fruition, comments computational biologist Philip M. Kim of the University of Toronto. For example, last year, Kim’s group designed 6,000 variants of an existing protein computationally and screened them to find nanomolar inhibitors of a target enzyme (Sci. Adv. 2016, DOI: 10.1126/sciadv.1600692).
The Baker study “is an important application of high-throughput mini protein design, selection, and production for discovering novel therapeutics,” says structural bioinformatics specialist Gaetano T. Montelione of Rutgers University. Structures of mini proteins bound to their targets could now be used for the rational design of even better small-molecule, macrocyclic, or optimized mini-protein drug candidates, he suggests.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter