Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
DNA
DNA nanostructure acts as lipid-flipping enzyme
Synthetic construct could someday replace damaged counterparts in disease
by Celia Henry Arnaud
July 2, 2018
| A version of this story appeared in
Volume 96, Issue 28

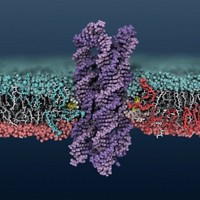
A DNA nanostructure flips lipids from one side of a lipid bilayer to the other more than 1,000 times as fast as natural enzymes called scramblases, researchers report (Nat. Commun. 2018, DOI: 10.1038/s41467-018-04821-5).
Asymmetry in the lipid composition of the inner and outer leaflets of biological membranes is essential to proper functioning of cells. To maintain this asymmetry, three classes of enzymes control movement of lipids between the inner and outer leaflets of membranes. One class, the scramblases, randomly moves lipids between layers to equilibrate the membrane composition, which can trigger cell death via apoptosis. Malfunctioning scramblases are associated with diseases such as Scott syndrome, in which blood doesn’t coagulate properly.

The DNA nanostructures reported by Ulrich F. Keyser of the University of Cambridge; Aleksei Aksimentiev of the University of Illinois, Urbana-Champaign; and coworkers act like synthetic scramblases. But that’s not what the researchers were originally looking for.
“We were interested in engineering DNA channels to act as artificial ion channels,” Aksimentiev says. They ran computer simulations of DNA nanostructures that Keyser’s group had previously made as ion channels. The nanostructures have eight DNA strands, two of which have cholesterol at one end to anchor the structure in the membrane.
Ion channels are supposed to make tight contact with a lipid membrane so nothing leaks across the barrier. But in computer simulations of their DNA channel, Aksimentiev’s team noticed lipids in the membrane were tilting, forming a toroidal pore around the DNA nanostructure that connects the two membrane layers. This pore was letting lipids along the outside of the nanostructure flip back and forth between the two sides of the membrane.
Experimentally confirming this scramblase activity took two years. But the researchers didn’t doubt they’d eventually succeed. “Once you see that the toroidal pore forms, the physics of the system says that it must scramble lipids,” Aksimentiev says, because the pore establishes a direct connection between the two leaflets.
To show the scrambling, Keyser’s group inserted the DNA nanostructures in synthetic vesicles made from a mixture of unlabeled and fluorescently labeled lipids. They induced bleaching of the fluorescence in the outer leaflet.
“If no scrambling happens, the lipids of the inner leaflet do not get bleached,” Alexander Ohmann, a graduate student in Keyser’s group, explains. “If we induce scrambling with our nanostructure, they would also get bleached.”
The researchers saw scrambling both in synthetic vesicles made from only one type of lipid and in human cancer cells with complex membrane compositions. Fluorescent microscopy measurements revealed that the nanostructure flips on the order of 10,000,000 lipids per second, which is three orders of magnitude more than natural scramblases.
“The comparison with an enzyme is correct,” says Friedrich Simmel, an expert in DNA nanotechnology at Technical University Munich. “Enzymes are catalysts that lower the activation barrier for a chemical process. But the case reported here is a very special enzyme: The energy barrier here really is the membrane structure, which prevents lipids flipping from one side to the other fast. By connecting the two membrane leaflets, the DNA structures simply remove this barrier.” Thus, although he considers the work to be a “remarkable achievement,” he doubts that it will be generalizable to other types of enzymes.
Advertisement
Right now the DNA nanostructures aren’t controllable, but the next step for Keyser’s and Aksimentiev’s teams is developing the DNA nanostructures so that they can be controlled by light or chemical stimuli. A longer-term goal is the possibility of turning them into drugs for Scott syndrome. “Rather than having a drug that repairs existing scramblases in the body, one could replace them with an artificial version built from DNA,” Keyser says. “That’s a very long-term goal, but our work shows that it could be possible.”
This article has been translated into Spanish by Divulgame.org and can be found here.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter