Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Mating mechanism diversifies synthetic libraries
Reshuffling library members can quickly lead to better binders of protein targets
by Celia Henry Arnaud
December 14, 2021

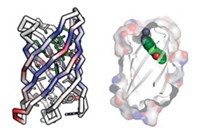
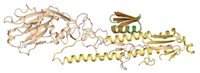
Using DNA to encode synthetic compound libraries makes it easy to identify library members with desired properties, such as affinity for a target molecule, using a screening assay. But the power of Darwinian selection hasn’t been harnessed for such synthetic libraries. Researchers have now come up with a “mating” mechanism for synthetic libraries that is conceptually similar to the recombination that happens in sexual reproduction (Nat. Chem. 2021, DOI: 10.1038/s41557-021-00829-5). The mechanism shuffles library members to generate new combinations not in the original library, and could help researchers find new inhibitors or drugs.
Nicolas Winssinger and coworkers at the University of Geneva used the mating mechanism to combine two smaller libraries into a diverse library of peptide assemblies that helped the researchers to identify candidates that bind a target protein—in this case, one important for understanding cancer. Following each round of selection, the best-performing library members can be broken into components that are copied and recombined to form new assemblies that weren’t present in the first round.
The libraries consist of peptides attached to DNA strands that both identify the peptide when sequenced and hold the peptide in a loop conformation. In this sense, the DNA serves as both a label and a structural component of the assembly. The researchers screen the library to find members that bind a target molecule with affinity above a certain threshold. The structural DNA strands associated with each loop in the selected assemblies can be copied separately and reshuffled to create new structures which undergo iterative rounds of selection until the assembly with the highest affinity is generated.
“The interesting thing of the mating mechanism is that the best molecule doesn’t need to be in the starting library to be selected through iterative rounds,” Winssinger says. “If the best molecule is made out of two loops that each contribute to the binding, as long as each of these loops is individually selected in the first round, it will be generated in a second or third round through this mating mechanism.”
The researchers demonstrated the use of their new approach by selecting library members that bind a protein called programmed death ligand 1 (PD-L1) and block its interaction with programmed death protein 1 (PD-1), a cell surface receptor on T cells that helps control immune responses. The geometry of the interaction between PD-L1 and PD-1 is such that it can be mimicked by the peptide loops in the library members.
Christian Heinis, a chemical biologist at the Swiss Federal Institute of Technology Lausanne who develops methods for synthesizing and screening large libraries of cyclic peptides, says it makes sense to apply such shuffling approaches to DNA-encoded libraries. “Although DNA encoding allows generating and screening large libraries of compounds (millions or even billions of compounds), the theoretical sequence space (all compounds that can in theory be generated combinatorially) is much larger, and it would never be possible to make and screen all of them,” Heinis writes in an email. “An approach in which new molecules are generated during the iterative selection process, using the ‘mating’ principle, is thus very interesting.”
Winssinger and his colleagues plan to also use the approach to make libraries of assemblies with peptide structures other than loops, including α-helices and coiled coils.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter