Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Gene Editing
2-in-1 base editors make two DNA edits at once
New dual base editors that can make A to G and C to T edits could have applications in plant science and synthetic biology
by Ryan Cross
June 3, 2020
| A version of this story appeared in
Volume 98, Issue 22

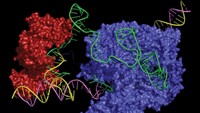
At least five groups have independently invented a new class of CRISPR gene editing tools called dual base editors. The systems combine the functions and components of two CRISPR base editors, which catalyze single nucleotide changes in DNA. The 2-in-1 tool is the latest in a growing line of CRISPR gene editing variants that allow scientists to perform new genetic tricks, or perform old tricks more smoothly.
David Liu’s lab at Broad Institute of MIT and Harvard developed the original base editors. His cytidine base editor (also called a cytosine base editor), first described in 2016, uses an enzyme called cytidine deaminase to ultimately convert a C into a T in DNA. Liu’s adenine base editor, published in 2017, uses an engineered bacterial enzyme called adenosine deaminase to kick-start a reaction with the end result of changing an A to a G. Both systems rely on a modified version of a CRISPR enzyme called Cas9 to target the base editor to a particular location in the genome.
To create the new dual base editors, scientists attached both the cytidine deaminase and the adenosine deaminase to Cas9. By using an easily programmable guide RNA to direct Cas9 to a desired stretch of DNA, scientists can use base editors to turn any A to a G and any C to a T in a narrow window.
A group led by Caixia Gao, a plant geneticist at the Institute of Genetics and Developmental Biology in Beijing, which is part of the Chinese Academy of Sciences, published the first peer-reviewed invention of a dual base editor in January (Nat. Biotechnol. 2020, DOI: 10.1038/s41587-019-0393-7).
Three more groups, one led by Nozomu Yachie at the University of Tokyo, another led by Dali Li at East China Normal University in Shanghai, and the third led by J. Keith Joung at Harvard Medical School, have described the creation of their own dual base editors in three papers published simultaneously this week (Nat. Biotechnol. 2020, DOI: 10.1038/s41587-020-0509-0; 10.1038/s41587-020-0527-y; 10.1038/s41587-020-0535-y).
And Beam Therapeutics, a public biotech company that is developing gene therapies derived from Liu’s base editors, filed a patent last year describing the design of dual base editors. That patent was published in February.
Although the groups took similar approaches to constructing dual base editors, there are slight differences in how each team attached the cytidine deaminase and adenosine deaminase to Cas9. Li says that although the idea of his group’s dual base editor wasn’t hard to come up with, it was not easy to make. “It took us much work to optimize the constructs for better performance,” he says.
Liu, who was not involved in the newly published studies, calls dual base editors a “clever, forward-thinking idea.” In applications where two kinds of DNA edits are needed, it may be easier to use a dual base editor than to attempt sequential editing with adenine and cytidine base editors, he adds.
Scientists can also use dual base editors to make new changes to codons—the trios of nucleotides that cellular machinery read when stringing together amino acids into proteins. “There are new amino acid substitutions that you can induce with the dual editors that you can’t induce with single base editors,” says Alexis Komor, a chemical biologist at the University of California San Diego who developed the first cytidine base editor as a postdoc in Liu’s lab.
Yachie says the dual base editors could be useful in human gene editing therapies, agriculture, and basic biology research. Two of the other groups have already put their editors to the test.
Gao’s lab developed several versions of dual base editors and tested them in rice cells. Crop scientists have long used chemicals and ultraviolet light to create random mutations in plants, with the hope that a mutation will improve specific traits of one lucky plant. Gao refined this idea by using dual base editors for targeted mutagenesis in rice cells. She let her base editors make random A to G and C to T edits in a region of a gene that makes rice susceptible to herbicides. By repeating this technique in a directed evolution experiment, Gao’s lab created herbicide resistant rice plants.
Li’s group provided a demonstration of how dual base editing might be used therapeutically. His lab used their editor to target a stretch of DNA that controls the expression of the γ-globin gene. γ-globin is a component of fetal hemoglobin, and its gene is normally turned off in adults. Reactivating the gene could provide a cure for people with genetic blood diseases like sickle cell disease. In human cell lines, Li’s dual base editor changed a CCA sequence, which repressed γ-globin production, into a TTG sequence, which promoted γ-globin production.
The dual base editors are an “important supplement” to the base editing toolkit, says Zhen Liu, who studies gene editing in animals at the State Key Laboratory of Neuroscience, part of the Chinese Academy of Sciences. Zhen Liu is “optimistic for the clinical application of the base editors,” including both single edit base editors and the new dual base editors.
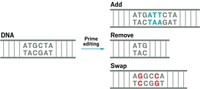
Several scientists say that dual base editors will need refinement before being used in humans. Others wonder how useful dual base editors will be for therapeutic purposes. Another kind of CRISPR gene editing developed by David Liu’s lab last fall, called prime editing, can in theory change one short sequence of DNA into any sequence, possibly negating and overshadowing the usefulness of the dual base editor’s A to G and C to T editing.
“It’s true that prime editors can make these changes as well,” David Liu says. However, the new editions of adenine and cytidine base editors are more efficient than their predecessors and the prime editor.
Komor envisions that plant scientists and synthetic biologists will find many uses for dual base editors in targeted mutagenesis and directed evolution experiments. She says that the number of instances in which it could be useful for human gene therapy could be much smaller.
Scientists at Beam, which is developing human gene therapies with adenine and cytidine base editors, agree. In a statement, Beam’s president and chief scientific officer Giuseppe Ciaramella says that dual base editors may have “limited utility” over single base editors because there are few instances where simultaneously changing an A to G and a C to T would be needed to treat a human disease. During the nearly two years that Beam has had dual base editors on hand, “we have not found attractive therapeutic options that could be uniquely or preferentially addressed by this technology,” he says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter