Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Gene Editing
David Liu unveils a search-and-replace CRISPR tool and a start-up to commercialize it
Beam Therapeutics and a new start-up called Prime Medicine will develop therapies based on prime editors, which can add, remove, or change nucleotides anywhere in the genome
by Ryan Cross
October 22, 2019
| A version of this story appeared in
Volume 97, Issue 42

Scientists just got a lot closer to being able to change DNA in any way they want thanks to a new CRISPR gene-editing system invented in David Liu’s lab at the Broad Institute of MIT & Harvard. The tools are called prime editors, and they might be the search-and-replace gene editors that biologists have been waiting for.
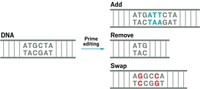
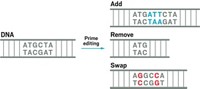
Contrary to popular belief, CRISPR-Cas9 gene editing can’t do it all. This classic CRISPR tool is great at making breaks in DNA, and biotech companies are already using it in experimental therapies to introduce mutations that turn genes on or off. CRISPR base editors, which were first developed by Liu’s lab in 2016, can change particular letters of DNA into other letters. Now Liu and a postdoc in his lab named Andrew Anzalone are debuting a prime editors, which can target any letter of DNA and turn it into any other letter, as well as insert new stretches of DNA or remove unwanted segments.
“If CRISPR-Cas9 and other programmable nucleases are like scissors, and if base editors are like pencils, then you can think of prime editors to be like word processors,” Liu says in a call with reporters.
Liu’s team has used his prime editors to make more than 175 edits in human cells in his lab, and he estimates that prime editors could be used to fix 89% of the 75,000 genetic mutations known to cause disease in humans (Nature 2019, DOI: 10.1038/s41586-019-1711-4).
Although prime editors have yet to be tested in an animal, and the technology, like previous CRISPR systems, has limits, it has attracted investors. Liu has cofounded a new start-up called Prime Medicine—a move that bolsters Liu’s status as a serial entrepreneur and underscores investors’ thirst for new gene editing tools.
Liu has previously cofounded two start-ups—Pairwise Plants and Beam Therapeutics—that use his base editors for agriculture and human therapies. Beam, which launched with $87 million in May 2018, recently filed paperwork for a $100 million initial public offering on Nasdaq. He insists there will be room for CRISPR-Cas9, base editors, and prime editors in the clinic.
Maria Jasin, a gene-editing pioneer and DNA repair expert at Memorial Sloan Kettering Cancer Center, says that prime editing could potentially overtake other gene-editing systems. “It’s difficult to tell from one paper, but it will certainly generate a lot of interest with people wanting to replicate it.”
Existing CRISPR systems have clear limitations. CRISPR-Cas9 cuts both strands of DNA, and when the cell tries to repair this break, it sometimes introduces a mutation by adding or removing a single letter of DNA, or nucleotide. That makes it good for turning problematic genes off or for turning silent genes on by disrupting their off switches. Many groups have tried inserting a new piece of DNA into that break as well, but the outcomes are inconsistent.
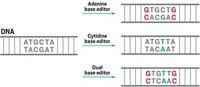
Cytidine base editors can convert C to T and G to A. And adenine base editors can convert A to G and T to C. But there are 8 other nucleotide changes—such as T to A and A to T—that these two classes of base editors can’t make. This limitation inspired Anzalone, who joined Liu’s lab in 2018, to devise a single system that could make any of the 12 possible nucleotide-to-nucleotide changes.
Anzalone created a molecular machine inspired by the classic CRISPR system, which uses Cas9 to cut DNA and a molecule called a guide RNA to tell Cas9 where to cut. Anzalone’s prime editor uses a specialized guide RNA to direct a modified version of Cas9 that makes a cut in just one strand of DNA’s double helix. That causes part of the DNA strand to flap out away from the helix. The flap serves as a DNA primer and gives prime editing its name. Next, the enzyme reverse transcriptase grabs the primer and uses it to begin writing a new sequence of DNA into the nicked site, looking to the guide RNA sequence as a template. Finally, the cell’s repair machinery fully incorporates the new DNA into both strands of the double helix.
The team tested multiple iterations of their editors in human and mouse cells. In one experiment, they used the technique to fix a single nucleotide mutation that causes sickle cell anemia. In other experiments, they used prime editors to remove 4 extraneous nucleotides that cause Tay-Sachs disease and added 3 nucleotides missing in a mutation that causes cystic fibrosis. Overall, Liu’s lab showed that prime editors could make all 12 single nucleotide changes, insert new DNA sequences up to 44 nucleotides long, and delete sequences up to 80 base pairs long.
Liu has made his prime editors available to use for noncommercial purposes from the nonprofit gene repository Addgene.
Prime Medicine, Liu’s new company, has already sublicensed the use of prime editors to his base-editor company Beam Therapeutics for certain applications. Beam’s website indicates that it is developing a therapy to fix a common sickle cell disease mutation that can only be corrected with prime editors.
“Prime editors will likely be more straightforward and widespread in their use” compared with base editors, predicts Ross Wilson, a CRISPR scientist at the University of California, Berkeley. Still, prime editors have limitations of their own. “Prime editors may be especially challenging to deliver because of their increased size,” he notes. Liu’s lab used an electrical zap to get DNA instructions for making prime editors into cells grown in a dish. That method won’t work for getting prime editors into animals or humans—a delivery job that will likely require packaging them inside nanoparticles or viruses. This is difficult to do with the classic CRISPR-Cas9, let alone the even bulkier prime editors, which include reverse transcriptase attached to Cas9, plus the guide RNA.
Liu also acknowledges that for now, his prime editors aren’t as efficient as existing base editors, and they still make mistakes—something that will likely need to be improved if they are used in humans. And then there’s the 11% of mutations that prime editors probably can’t tackle, such as extra or missing copies of entire genes—which are likely too large for prime editors to remove or add.
“Prime editing will not replace base editing or other CRISPR-Cas9 tools,” says Chloe Christensen, a graduate student in Francis Choy’s lab at the University of Victoria, who is developing base editors to fix mutations that cause lysosomal storage diseases. Nonetheless, Christensen says that prime editors “eliminate the rigid parameters” of the existing base editors. “If Cas9 is scissors and base editors are tweezers, prime editors are the Swiss Army Knives, the ultimate multi-tool.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter