Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Natural Products
Biosynthesis of eagle-killing toxin elucidated
Pathway includes unusual enzymes including a single-component flavin-dependent halogenase
by Celia Henry Arnaud
February 18, 2022

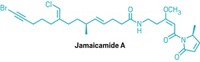
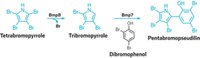
Researchers have determined the biosynthetic pathway for a cyanobacterial toxin known as aetokthonotoxin (AETX). The toxin is highly brominated, which is rare in both cyanobacteria and freshwater environments.
In 1994, scientists discovered a neurological disease in bald eagles that causes lesions on the brain and spinal cord. It took more than a quarter century for researchers to connect the disease to AETX, which is produced by a cyanobacterium that grows on the invasive aquatic plant Hydrilla verticillata (Science 2021, DOI: 10.1126/science.aax9050). Although they identified the gene cluster responsible for the synthesis, they didn’t elucidate the pathway.
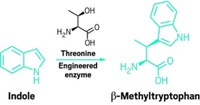
Bradley S. Moore and coworkers at the University of California San Diego have studied polybrominated natural products in saltwater environments. They were intrigued by AETX, which has five bromines and a nitrile, and wanted to figure out how it’s made.
They found that AETX comes from two tryptophan building blocks that are separately decorated before being coupled (J. Am. Chem. Soc. 2022, DOI: 10.1021/jacs.1c12778). A flavin-dependent halogenase (AetF) adds bromine to tryptophan as the first step in both branches of the pathway. In one branch of the pathway, AetF adds a second bromine, and then another enzyme called AetD converts tryptophan’s alanyl side chain to a nitrile. In the other branch, enzymes remove the alanyl group and add two bromines. Finally, a fifth distinct enzyme couples the two indoles in the modified tryptophans.
AetF and AetD are both unusual enzymes. Most flavin-dependent halogenases work with a partner protein for cofactor regeneration, but AetF “doesn’t need a partner because it has that reductase capacity within the same protein,” Moore says. That could make AetF attractive for industry as a biocatalyst. “If you can have one enzyme versus two enzymes, it’s so much simpler,” Moore says.
AetD has left Moore and his colleagues trying to figure out how one enzyme can convert an alanyl group to a nitrile. “We still need to learn more about how that enzyme ticks and what it’s able to do,” Moore says. “It’s doing a lot more than a synthetic chemist would be able to do in a chemical reaction with a single reaction step.”
“AetF is very interesting since it would eliminate the need to add a separate reductase for cofactor regeneration,” Jared C. Lewis, a chemistry professor at Indiana University Bloomington who studies biocatalysis, writes in an email. “I was most intrigued by the AetD-catalyzed nitrile formation. That could be useful for biocatalysis, but the enzyme that catalyzes it will almost certainly be interesting simply due to the nature of the bonds that must be formed and broken in that transformation.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter