Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Natural Products
Cultured bacteria make hypermodified peptides
Engineered plasmids could lead to new sources of drugs
by Celia Henry Arnaud
September 12, 2019
| A version of this story appeared in
Volume 97, Issue 36

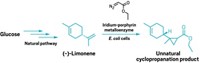
Polytheonamides are extensively modified peptides produced by bacteria in the microbiomes of marine sponges. These pore-forming peptides are toxic to cells, which makes them intriguing as possible cancer drugs. A major problem is that the bacteria that make the peptides can’t be cultured.
Now, Jörn Piel of the Swiss Federal Institute of Technology (ETH), Zurich, and coworkers have identified a cluster of enzymes in another bacterium that produces similarly hypermodified peptides (Nat. Chem. 2019, DOI: 10.1038/s41557-019-0323-9). And unlike the bacteria from the marine sponge, Microvirgula aerodenitrificans can be grown in cell culture.
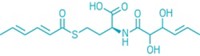
M. aerodenitrificans produces its own class of modified peptides called aeronamides. The peptides are ribosomally synthesized with leader and core portions. The enzyme cluster recognizes the leader portion and modifies the core. The modifications include iterative epimerization to introduce 21

The yield isn’t very good at this point, however. After growing the bacteria for 2 days in a 5 L cell culture, the researchers were able to isolate only about 600 µg of the peptide aeronamide A. “We’re working on improving promoter systems and trying a different plasmid so that we can up the production of the peptide,” says Agneya Bhushan, a graduate student in Piel’s lab who did much of the work.
The researchers “have a way to go before they are able to robustly produce material,” says Bradley S. Moore, a natural products expert at the University of California San Diego. “But given the significant structural complexity, that result is quite remarkable and a tremendous first step that gave them insight into the potent bioactivity” of aeronamide A.
The researchers are now trying to engineer other core sequences that can be similarly modified, and they plan to screen such peptides against pharmaceutical targets.
CORRECTION
This story was updated on Sept. 17, 2019, to correct the location of one of the C-methylations in the peptide structure.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter