Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Proteomics
An abundance of allostery found by combining biochemical assays and AI
Technique combines lab assays and computer algorithms to hunt out proteins’ hidden pockets
by Laura Howes
April 14, 2022
| A version of this story appeared in
Volume 100, Issue 13
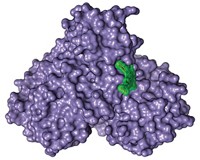
Researchers at the Centre for Genomic Regulation have developed a new technique for identifying allosteric sites in proteins (Nature 2022, DOI: 10.1038/s41586-022-04586-4).
While many drugs target the active site of a protein, where the chemistry happens, others take a more sideways approach. Allosteric sites are additional pockets that drugmakers can use to regulate the activity of enzymes, dialing their activity up or down by shifting the shape of the protein. Allosteric inhibitors can also be used to target disease-causing mutant proteins, such as those made by cancer cells, without affecting the analogous healthy proteins.
The team, led by Ben Lehner, analyzed two well-characterized human protein–binding domains and found many more sites with allosteric potential than had previously been identified. The researchers say the new technique could be a boon for drug discovery.
Until recently, finding allosteric sites has been expensive and time consuming. In their new work, the researchers used common cell assays that measure protein binding or abundance. In these assays, if the enzyme is functional, the cells will gain resistance to an antibiotic, meaning cells’ growth rate in the presence of the antibiotic is a proxy that lets scientists quantify protein binding. The researchers made thousands of slightly different versions of a protein by systematically mutating it; they then ran assays on all versions of the protein. By feeding all their assay data into a neural net, the team could pinpoint potential allosteric sites, characterize them, and explore how they could be modified.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter