Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Proteomics
Many of our proteins remain hidden in the dark proteome
Scientists want to develop new tools and methods to uncover the proteins they’ve yet to see
by Laura Howes
January 24, 2022
| A version of this story appeared in
Volume 100, Issue 3

Credit: Alfred Pasieka/Science Source, SPL/Science Source(Proteomics); Shutterstock (Face)
Every second of every day inside your body, proteins are made, moved, modified, and destroyed over and over again. These proteins can be short strings of amino acids or longer, intricately folded shapes. They can send messages or combine with other proteins to build machines or structures in the cell. Proteins are the performers that swirl and dance across every part of our bodies, and layers of complexity play out at the molecular level.
In brief
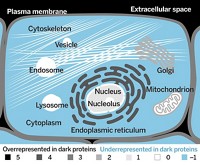
By some measures, scientists have nearly finished quantifying the human proteome. But by others, they have only just scratched the surface of the variety and complexity of our bodies’ proteins. They have yet to find proteins for some known genes, and they haven’t characterized the modified versions of some proteins. For other proteins, scientists don’t know what they look like or what they do in cells. Researchers say that uncovering this dark proteome is one of the significant challenges in biology and will affect how we understand our biochemistry and our health.
That dance matters. When outside dancers join or a key member of the ensemble becomes injured, the choreography changes. Those effects might be subtle but could have significant consequences for our health. It’s how tiny viruses can send people to the emergency room or small changes in our DNA can grow into cancers.
When it comes to the lives of proteins in our cells, there is a constant whirl of activity. But scientists may be seeing only a fraction of it. Researchers have trained their spotlight on many proteins in our cells, but other proteins perform their movements in the dark, out of scientists’ view.
These unknowns, both known and unknown, are the dark proteome. Inside this dark proteome are proteins that scientists think should exist but haven’t found, proteins that can be constructed and modified in different ways, and proteins that scientists have found but whose structures and roles are still unknown. Researchers hoping to learn more about our biology, including details about diseases and how to treat them, are developing new tools and ways to explore this dark proteome.
“For me,” says Sean O’Donoghue at the Garvan Institute of Medical Research, “darkness is a metaphor for what we don’t understand.”
The undetected
When researchers first announced the human genome sequence 20 years ago, they also estimated how many different proteins are in our cells. These scientists involved in the Human Genome Project looked for each bit of DNA that signals the start and end of a protein and counted 20,000 genes that code for proteins, about 1% of our DNA.
While some of these proteins have been easily detected, crystallized, and characterized, others have taken longer to detect. Different teams have been trying to fill in the gaps.
In 2014, two teams of researchers, one led by Akhilesh Pandey at the Johns Hopkins University School of Medicine and the other led by Bernhard Küster at the Technical University of Munich, each published draft maps of the human proteome after analyzing different human tissues and fluids by mass spectrometry. The two teams detected 80–90% of the proteins predicted to exist. A third effort, the Human Proteome Project (HPP), which involves researchers from around the world, has also sought evidence of proteins thought to be encoded in the human genome. The HPP says that there is evidence for just over 90% of those proteins.

That means that of the 20,000 discrete sections of our DNA that are thought to code for proteins, researchers have evidence for over 18,000. What remains is the first part of the dark proteome: the undetected.
“A whole bunch of them could be really important, and we’ve got no clue because we can’t even study them,” says Christopher Overall, who studies proteomics at the University of British Columbia.
Scientists haven’t detected those proteins for many reasons, Overall says. For one, cells might not be expressing the protein. Or maybe cells do express it but only at low amounts or at a particular time. Analytical techniques can provide only a snapshot of the proteins at work inside a tissue or body fluid at a given time. If the protein isn’t present when a sample is analyzed, scientists will keep missing it. One way around this problem is to look for the proteins in tissues that scientists don’t normally sample, such as olfactory and sperm cells.
But maybe the proteins are in samples but lack features that allow them to be detectable by current analytical techniques. For example, analyzing proteins by mass spectrometry requires first breaking them into smaller fragments, or peptides. To do this, scientists usually use a protein-chopping enzyme called trypsin. But if trypsin can’t digest a protein in the sample, the mass spectrometer won’t be able to analyze the molecule. Scientists can use other enzymes to digest protein samples for mass spectrometry, Overall says, but that requires a lot of effort to modify protocols and for not much reward, so researchers often opt to study less-fussy proteins.
According to the HPP, there are currently 1,421 “missing proteins”: those that are predicted to exist but haven’t been measured experimentally. The problem, Overall says, is that finding those missing proteins is becoming more and more difficult. The fussiest proteins need a lot of work to find. And while a new protein might ultimately be found to be critical to a cell or a target for disease treatment, the payoff isn’t guaranteed, so grants for looking for undetected proteins are scarce. That means scientists often run these search projects on the side.
But as researchers try to detect the missing proteins, there are still other regions of the dark proteome to explore.
The disguised
By one count, slightly under 10% of the proteins dancing in our cells are unknown to scientists. But from another perspective, about 90% of the proteome could be hidden—many proteins still dance in the dark. The difference in measuring the dark proteome depends on what it means to identify a protein.
The HPP determines that a protein is found when scientists have evidence of peptides that match the protein sequence predicted from a specific gene. But the truth is that there isn’t a one-to-one relationship between genes and proteins, according to Lydie Lane of the SIB Swiss Institute of Bioinformatics. Instead, each gene can be expressed in different ways, and the resulting proteins can also be modified after being produced.
When you account for those variations, there could be millions of varieties of proteins inside each of us, all coming from the same 20,000 genes, Lane says. That would massively multiply the complexity of the proteome. “We don’t have the means to know how many there are, and when and why, etc.,” she says. “So this is quite dark.”
At molecular biology’s simplest, genes in DNA are transcribed into RNA and then translated into proteins. But because biology loves to be complex, the protein-making process doesn’t always start at one end of a gene and then read through to the other end. The RNA that will be translated into a protein is often spliced together in different ways. This splicing means that while the gene might code for protein sections 1, 2, 3, and 4, the spliced RNA could tell the ribosome to make a protein consisting of 1-2-3-4 or 1-3-4 or 1-2-4, or it could even skip bits of one of the sections. These different forms of the expressed gene are called isoforms. Each resulting protein comes from the same gene and could look the same when digested and fed through a mass spectrometer, but the different versions of the molecule could have different roles in the body. For example, scientists have found 12 isoforms of the enzyme 5′ adenosine monophosphate–activated protein kinase. While they all help regulate cellular energy, they all do slightly different things in different places in the body.
One gene, many proteins


To make the protein picture even more complex, different machinery within a cell can chemically modify proteins. These so-called posttranslational modifications can be small additions like methyl or acetyl groups or larger ones such as sugar molecules. For example, in acute pancreatitis, the nitration of an enzyme called cystathionine β-synthase reduces its activity, which causes rapid reductions in key biomolecules. Another way proteins can be modified after synthesis is proteolytic processing, in which enzymes snip proteins into shorter molecules for other roles. For example, a longer protein gets cut down into the biologically active form of the hormone insulin.
Together, all these variations to proteins could result in potentially millions of what are called proteoforms.
“It’s something that people have known is an important part of biology forever,” says Parag Mallick, who uses proteomics to look for cancer biomarkers at Stanford University. “We just didn’t have tools that could actually measure them.”
Mallick cofounded a company, Nautilus Biotechnology, to commercialize his idea for a more sensitive method of protein detection and identification. The common mass spectrometry approach to identifying proteins is destructive, which means scientists have just one shot at looking for what is in a sample. Scientists can easily miss subtle variations in a protein’s composition and not have a second chance to catch them. The Nautilus system immobilizes proteins from a sample onto a surface and then uses fluorescent reagents that can bind to specific protein structures and modifications to repeatedly probe what’s there. By applying different probes to the immobilized proteins, the scientists hope to get a truly representative readout of the proteins in a sample (bioRxiv 2021, DOI: 10.1101/2021.10.11.463967). This paper is a preprint and has not yet been peer-reviewed.
Nautilus went public in June 2021, raising $345 million for the company, which had investors such as Amazon snap up shares. The firm’s technology is available only to current research partners, but Mallick hopes it will become available commercially in the future. Other groups are also developing new protein-sequencing technologies. For example, some researchers use a technique in which they pull a protein through small openings called nanopores and use changes in electrical conductivity across the pore to read the amino acid sequence and keep track of the protein’s chemical modifications.
The shapeless
But while a protein’s amino acid sequence and posttranslational modifications can tell researchers a lot about the molecule, proteins don’t just exist as a linear chain of building blocks. Proteins do their actions mostly when their chains fold up correctly.
And that is another dark area of the proteome—proteins with unknown structures or functions. For these proteins, scientists don’t know what the molecules look like, which other molecules they interact with, or even what they do in our bodies.
Recent developments in artificial intelligence–based methods have helped researchers start to predict structures for some of these dark proteins (Nature 2021, DOI: 10.1038/s41586-021-03828-1). These algorithms take the amino acid sequences of structurally dark proteins and then use what they’ve learned from data on known protein structures to predict how the dark ones might fold. These predictions can help researchers when experimentalists haven’t captured the 3D form of a protein. But the truth is, not all proteins have fixed structures.


Protein structures are constantly moving and adjusting. And sometimes proteins don’t adopt a defined 3D structure to begin with. Structural biology techniques like cryo-electron microscopy and X-ray crystallography provide only a snapshot of what a protein looks like. Some proteins might have a given structure only in specific environments, and some proteins steadfastly refuse to hold a 3D shape for experimentalists to capture. The latter type of protein includes intrinsically disordered proteins, which continually wiggle and wobble about, and membrane proteins, which are notoriously difficult to crystallize. So does the structurally dark proteome comprise just intrinsically disordered proteins? No, says O’Donoghue from the Garvan Institute.
Back in 2015, O’Donoghue and colleagues investigated the structurally dark proteome. “The shock to us,” he says, “was that most of the darkness was not disordered.” Instead, he found that this group of proteins was pretty heterogeneous. There certainly were some disordered proteins, but there were also ordered ones. So while the proteins have properties that preclude capturing these structures with current techniques, O’Donoghue suspects that some do have defined structures and perhaps fold in ways that researchers haven’t yet seen.
“My suspicion is that there are probably categories of folds and probably certain kinds of transmembrane folds, for example, that are just fundamentally different to what we know,” he says. Transmembrane regions are often strongly hydrophobic, making them hard to dissolve and handle for standard structural biology techniques. If these proteins have structural features that haven’t been seen before, AI algorithms trained on existing knowledge might struggle to make accurate predictions about these dark proteins. But researchers are confident that they can start to shine light on this darkness with a combination of improved structural techniques and the growing power of prediction algorithms.
One reason why scientists want to solve the structures of these dark proteins is that the structure often helps point to a protein’s functions. Structure and function are inevitably linked, says Burkhard Rost at the Technical University of Munich. “The fact is we cannot predict pathways from structure,” he says, “but without structure, important aspects of pathways remain unclear.”
Advertisement
Rost himself moved from solving protein structures to discovering proteins’ function. He says that the functionally dark proteome is much harder to define than the structural one. It is much more of an unknown unknown, he said.
“It’s a bit weird and a bit disturbing,” agrees Lane of the Swiss Institute of Bioinformatics. There are some proteins, she says, that “are super abundant. We know them. They are validated. You can measure them, not only in human but also in other species. But you don’t have any clue about their function.” She’s been building a database of protein function to try to prompt people to fill in the gaps.
Validating the function can involve experimental techniques such as silencing a specific gene in a cell to see what happens in the protein’s absence. Rost is trying AI approaches. For example, he recently led a team that used an AI algorithm called Deep Learning to predict if specific stretches of proteins could bind things like small molecules or metals (Sci. Rep. 2021, DOI: 10.1038/s41598-021-03431-4). Knowing what a protein interacts with can give scientists a clue about its function. But that work took several years and uncovered only a few previously unknown binding domains.
“There is a lot of darkness, if you call darkness the lack of functional annotation,” he says. “And this darkness is huge.” While AI might have helped illuminate large parts of the dark structural proteome, he adds, researchers still haven’t built a way of defining function that would allow deep-learning algorithms to solve the puzzle, and the data that exist are too sparse for algorithms to learn from. Bioinformatic databases such as the Gene Ontology resource collect information on gene functions, he says, but they aren’t far reaching enough, especially because proteins can have multiple roles depending on their biological context. So scientists will need to do more work to better characterize protein function before AI can help explore this dark region of the proteome.

Over the past hundred years, scientists have uncovered a huge amount of information about the community of proteins inside our bodies. But each stage has shown that scientists had a lot more to learn. What’s more, these proteins living in the dark proteome might be just the beginning.
There may also be other areas of darkness that scientists don’t even know about yet. For example, Lane says, perhaps there is an unpredicted proteome—pieces of the genome that researchers don’t realize are genes but that encode proteins or small peptides. “There are probably even darker things,” she adds. “It’s difficult to know what we don’t know.”
One thing everyone can agree on is that just because a protein is hidden doesn’t mean it is unimportant. Researchers like Mallick think new technologies will help shed light on these dark areas of the proteome. Put simply, he says, “if there’s stuff that you’re not seeing, and it’s part of what’s driving the behavior of a system, you want to see it.” And that’s why the mysteries of the dark proteome are going to hold researchers’ attention for a while to come.
CORRECTION
This story was updated on Jan. 27, 2022, to correct the description of the availability of Nautilus Biotechnology's protein analysis technology. The companies that have access to the technology are better described as Nautilus’s research partners, not commercial partners. The company hopes to make the technology commercially available in the future.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter