Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein Racemate Yields Crystals
Racemic crystallography gives structures of uncrystallizable proteins
by Amanda Yarnell
January 26, 2009
| A version of this story appeared in
Volume 87, Issue 4

NEW WORK SUGGESTS that a technique called racemic crystallography could help structural biologists get high-quality protein crystals of tough-to-crystallize proteins.
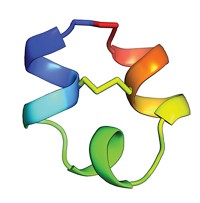
Chemistry professor Stephen B. H. Kent, postdoc Kalyaneswar Mandal, and coworkers at the University of Chicago show that they can readily obtain high-quality crystals of the protein BmBKTx1 from a chemically prepared racemic solution (J. Am. Chem. Soc., DOI: 10.1021/ja8077973). Found in the venom of a scorpion native to Asia, the 31-residue protein toxin is a poster child of recalcitrance, having resisted extensive crystallization attempts by multiple labs.
The idea that a racemic protein mixture would crystallize more readily than either enantiomer alone first came from UCLA structural biologist Todd O. Yeates back in 1995. The Kent lab's work "has borne out that prediction," Yeates says.
The BmBKTx1 structure is one of only a handful of protein structures ever determined by racemic crystallography. The method has languished in relative obscurity, Yeates notes, because it requires that proteins made purely of D-amino acids be created to mirror conventional L-amino acid-containing proteins. D-Amino acid proteins cannot be made by the usual recombinant methods; rather, they must be chemically synthesized. "The perceived difficulty of chemical synthesis of proteins has stood in the way," Kent says.
"But protein synthetic methods have become increasingly powerful," notes Jeremy M. Berg, who published the first example of racemic protein crystallography in 1993 and is now the director of NIH's National Institute of General Medical Sciences. Kent says his lab can now chemically synthesize proteins of up to 300 amino acids rapidly and in high purity.
Racemic crystallography is likely to be a general approach to getting structures of difficult-to-crystallize proteins, predicts crystallographer Ted Baker of the University of Auckland, in New Zealand. Baker's group teamed up with the Kent lab to try the method after repeatedly failing to crystallize a 94-residue protein made by the bacterium that causes tuberculosis. It "provided us with our only route to crystals," he says.
The method is "probably not yet applicable to structural genomics, where the aim is to speed up, streamline, and automate each step," Baker says. Kent disagrees, arguing that "the era of rapid total synthesis of proteins is at hand."


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter