Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Pharmaceuticals
From Picture To Pill
Debut of G-protein-coupled receptors will bolster drugmakers’ tool kits
by Carmen Drahl
March 14, 2011
| A version of this story appeared in
Volume 89, Issue 11

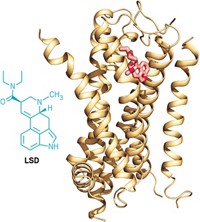
By the time Sid Topiol ended up at Novartis, he’d been thinking about G-protein-coupled receptors (GPCRs) for quite some time. A computational chemist turned molecular pharmacologist, Topiol had often encountered the membrane-straddling receptors on his research path. Yet as late as 2000, he knew little about what they looked like. “I remember the time when a GPCR was something you envisioned as just a big blob,” he says. Researchers knew GPCRs snaked back and forth across the cell membrane seven times. But that wasn’t much help for drug hunters, for whom the finer points of protein structure are paramount.
Then, on Aug. 4, 2000, structural biologists published the first X-ray structure of a GPCR. That day, an elated Topiol charged into a meeting, the journal article held aloft. “It was all very exciting,” recalls Topiol, now chief scientific officer at 3D-2Drug, a firm that facilitates collaborations for structure-based drug discovery. But in retrospect, “it was almost like a tease.”
That’s because rhodopsin, the first GPCR to have its structure determined, isn’t like most GPCRs. It’s more stable than most others outside the cell membrane, and it responds to light, not to a molecule, like most other GPCRs do.
It took seven years before the next GPCR structure—that of the β2-adrenergic receptor, which responds to adrenaline—was solved by two teams, Brian K. Kobilka’s at Stanford University and Raymond C. Stevens’ at Scripps Research Institute (Nature, DOI: 10.1038/nature06325; Science, DOI: 10.1126/science.1150577). In the past few years, the trickle of new structures has started to gain speed. So far structures of six GPCRs have been published, with more on the way.
Researchers are excited about the influx of GPCR X-ray structures because of their potential to aid drug discovery. As many as 40% of marketed drugs, including Bristol-Myers Squibb and Sanofi-Aventis’ blockbuster blood thinner Plavix and Pfizer’s allergy treatment Zyrtec, control these signaling proteins. Because GPCRs have a hand in so much of what the body does, companies would like to get even better at bringing drugs that act on them to market.
The new structures aren’t a panacea, and translating the information they hold isn’t without obstacles. But in the arduous quest that is drug discovery, researchers say, every piece of information helps.
Historically, drugmakers have been very successful at finding drugs that act on GPCRs, says Xiayang Qiu, senior director and head of Pfizer’s structural biology and biophysics group. “But the low-hanging fruit has already been picked.”
The relatively easy pickings were GPCRs that evolved to respond to compact, druglike molecules such as dopamine, Qiu says. But selectivity among multiple subtypes of a GPCR remains a challenge. And that’s not the hardest problem, he adds. “For some GPCRs, the natural ligand is not druglike, or we don’t even know what the ligand is.”
Many large companies are excited at the prospect of using structural information to tackle those more complex problems, Qiu says. For instance, the glucagon-like peptide-1 (GLP-1) receptor, the target of injectable diabetes treatments such as Eli Lilly & Co. and Amylin Pharmaceuticals’ Byetta, has a non-druglike ligand. Oral GLP-1-targeted drugs would be easier than nonoral agents for diabetes patients to take, but none has been developed yet. GLP-1 responds to a peptide, which means finding an oral drug “is really a chemistry issue—to identify the small molecule that interacts in just the right spot to get the same level of efficacy” as a peptide, Qiu says.
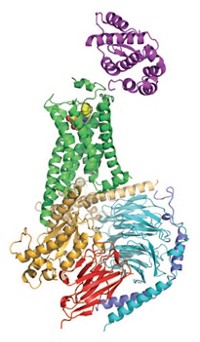
Smaller companies are intrigued by GPCR structures as well. Researchers at Cambridge, Mass., biotech outfit Anchor Therapeutics would like to obtain a GPCR X-ray structure that can help them learn more about out how their pepducin technology works. The company makes lipopeptides called pepducins that control GPCR activity. One of its advanced pepducins activates C-X-C chemokine receptor type 4 (CXCR4), a GPCR that helps recruit stem cells to sites of injury. The X-ray structure of CXCR4 was reported for the first time last year (Science, DOI: 10.1126/science.1194396). The receptor activates two pathways, and the pepducin triggers only one of the two, making it what’s called a biased ligand. Biased ligands are a hot field in GPCR drug development because they have the potential to activate GPCR pathways associated with a desired pharmacological effect while having no effect on pathways associated with side effects (C&EN, Jan. 10, page 22).
Anchor scientists know that pepducins traverse the cell membrane, where they are thought to stabilize GPCR conformations. But knowledge of the exact GPCR conformation stabilized would go a long way toward helping them understand how biased ligands like pepducins work, and perhaps even make it easier to design biased ligands in general. “We’re looking to collaborate with labs that do structure-based work on CXCR4,” says Kenneth Carlson, Anchor’s vice president of biology. “A home run would be to have an X-ray structure of a GPCR target together with a pepducin.”
Though GPCR structural biology hasn’t yet tackled that problem, it is already helping chemists who seek drugs, from computer modelers to medicinal chemists.
Crystal structures provide the finest level of GPCR structural detail yet, Scripps’s Stevens says. They show “all the pockets on the protein, all the possible places you can go in terms of designing a molecule.”
“Every GPCR has seven helices. So you might think—how different can the pockets really be?” Stevens says. It turns out they’re quite different, he says. A GPCR’s seven helices form a protein cup to hold different molecules. “It’s similar to how many different types of drinking glasses exist.”
“We have pinot noir glasses, we have champagne flutes, and we have beer mugs. They all have one thing in common—glass formed in a specific shape holding different liquids,” Stevens says. But their functions are distinct.
When it comes to figuring out which nooks and crannies of a protein pocket to exploit, it’s been exciting to see structures of several distinct GPCRs as well as the same GPCR binding to different ligands, says Andrew J. Tebben, principal scientist in computer-aided drug design at Bristol-Myers Squibb. For instance, since 2007, multiple structures of the β2-adrenergic receptor have been solved, he says.
“The first structure obviously teaches you a lot,” Tebben says. But drug discovery teams tend to use multiple X-ray structures to do iterative design, in which a protein might be crystallized together with multiple different compounds under evaluation. This process can help chemists figure out the best spots to modify on a compound to improve one property, such as binding affinity, without affecting another, such as metabolic stability. For drug discovery on other protein targets, such as kinases, this so-called iterative design is the norm, but for most GPCR targets the technology hasn’t matured to the point that iterative design is possible yet, Tebben says. “I think that’s going to change pretty dramatically in the next few years,” he says.
GPCR crystal structures are already changing the ways medicinal chemists think about drug design, says Amy H. Newman, chief of the Medicinal Chemistry Section at the National Institute on Drug Abuse. Until recently, “we never expected that a [GPCR] crystal structure would be available for us,” so medicinal chemistry for GPCRs used to focus not on the protein structure but on the small molecules themselves, she says. For example, medicinal chemists might make a series of modifications on a dopamine receptor blocker and then test the series in biochemical assays to determine how structural changes affect binding affinity to the dopamine receptor and dopamine receptor function.

That change-and-check approach has been reasonably successful—her team has discovered selective blockers of the dopamine D3 receptor that are being tested in animal models of drug addiction. “But are they all perfectly druglike? No,” Newman says. Now that her team has obtained an X-ray crystal structure of the dopamine D3 receptor, published last year in collaboration with the Stevens team, they can make compounds that might fit the binding site more closely (Science, DOI: 10.1126/ science.1197410).
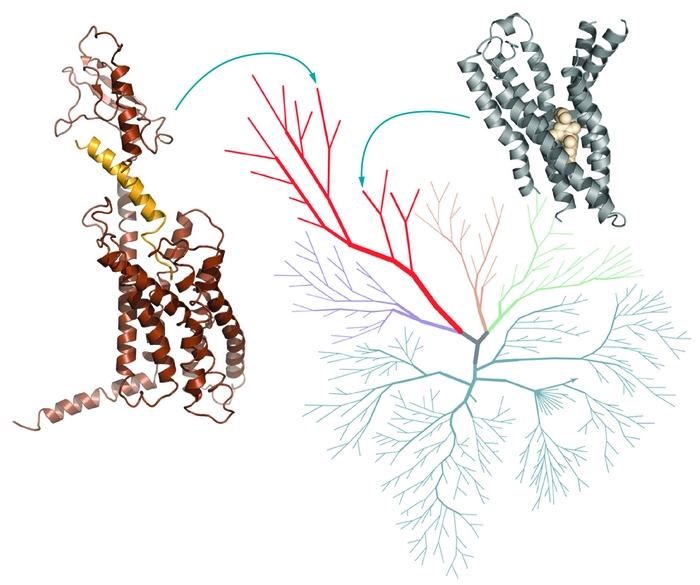
Every new GPCR structure makes computer models a little bit more reliable, and as a result a little bit more useful for predicting unknown GPCR structures or hunting for small molecules that act on GPCRs, says Ryan G. Coleman, a postdoctoral fellow in Brian K. Shoichet’s computational chemistry group at the University of California, San Francisco. Coleman interned at a pharmaceutical company in the early 2000s, a time when researchers were using the published X-ray structure of rhodopsin to create homology models, three-dimensional best guesses about what related GPCRs would look like. Because rhodopsin was so different from most GPCR drug targets, “people would make models, but it was still hard to believe them,” Coleman says. With more structures available, more reliable models will follow, he says.
Those models are useful for predicting related GPCR structures and for predicting how different states of a GPCR might look, says Vsevolod Katritch, a project scientist in Ruben Abagyan’s computational chemistry group at the University of California, San Diego. GPCRs have different conformations in their “off” and “on” states. But until last Januray, all published X-ray structures of clinically relevant GPCRs were of inactive states. And it wasn’t clear how useful those structures would be for discovering drugs that turn GPCRs on instead of turning them off.
Katritch and colleagues in the Abagyan and Stevens groups used modeling to predict interactions of small-molecule activators binding to the β2-adrenergic receptor and the A2A receptor, a GPCR that responds to adenosine (J. Mol. Recognit., DOI: 10.1002/jmr.949; J. Biol. Chem., DOI: 10.1074/jbc.M109.096974). Those predictions, including some conformational changes in the small-molecule binding pocket, were borne out in subsequently published X-ray structures of the proteins bound to activators (Nature, DOI: 10.1038/nature09648, DOI: 10.1038/nature09746, and DOI: 10.1038/nature09665), including specific active-state structure of the A2A receptor bound to an activator published just last week (Science, DOI: 10.1126/science.1202793).
The new GPCR structures aren’t only useful for modeling other protein structures—they can also help researchers screen potential drug molecules, Shoichet says. Typically, companies begin the search for molecules that control the activity of promising GPCR targets with fluorescent assays of library compounds. But it’s not always cost- or time-efficient to screen every single molecule in a vast company library. So teams can use GPCR X-ray structures to virtually screen vast numbers of molecules in order to eliminate the least likely possibilities.
Virtual screens can turn up hits that look different from those found in conventional assays, Shoichet adds. Such new chemical scaffolds are exciting because they represent new ways of controlling a GPCR’s activity and provide alternative chemistry solutions for problems that may come up during drug development, he explains. In his team’s virtual hunt for blockers of the A2A receptor, many of the best hits looked different from adenine and caffeine—two of GPCR’s best-known blockers (J. Med. Chem., DOI: 10.1021/jm100240h).
The modeling community has been using emerging GPCR structures in an assessment program called GPCR Dock, begun in 2008, to evaluate their progress and learn how to improve their models, says Irina Kufareva, a project scientist in the Abagyan group. Last summer, just before the release of new GPCR X-ray crystal structures of the dopamine D3 and CXCR4 receptors, researchers sent homology models of those proteins, with ligands bound inside, to GPCR Dock coordinators to see how close they’d come to the real thing. The teams’ predictions were more accurate for the dopamine receptor than for CXCR4. It wasn’t a surprising result, because CXCR4 is rather different from other GPCRs that have been analyzed by crystallography, and modeling works best when there’s a closely related template to work from, Kufareva says. She hopes the assessment will encourage modelers to explore new approaches and experimentalists to solve structures strategically—picking the most diverse GPCRs so that modeling will be better able to fill in knowledge gaps.
As helpful as GPCR X-ray structures are, they aren’t going to make all the challenges of drug discovery evaporate, and they aren’t going to replace established technologies so much as complement them. Structures are static snapshots of dynamic proteins. GPCR structures are still harder to come by than typical X-ray structures. And compared with established assays, they are of limited utility for the discovery of new allosteric drugs, an important class of GPCR-targeted drugs, some researchers say.
Advertisement
Allosteric drugs bind receptors at sites different from those to which natural ligands bind (C&EN, Nov. 23, 2009, page 12). But by and large, GPCR X-ray structures only include molecules that bind to sites where natural ligands bind, called orthosteric sites.
Despite their limitations, GPCR structures will be a valuable addition to the drug discovery tool kit, Stevens says. And if more X-ray structures can help drugmakers discover GPCR-targeted drugs—as they do for other targets, such as kinases—it’s a resounding validation of recent investments in basic research on GPCR structural biology by the National Institutes of Health Common Fund and the National Institute of General Medical Sciences’ Protein Structure Initiative.
“But is structure all you need? Absolutely not,” Stevens says. “You need to have a good target, you need to understand the biology, and you need solid medicinal chemistry,” he says. “The real magic will come in combining structure with all of these things.”




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter