Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Biochemistry: Proteins Containing Unnatural Amino Acids
Scientists and biotech companies advance efforts to incorporate unnatural amino acids into proteins
by Carmen Drahl
December 19, 2011
| A version of this story appeared in
Volume 89, Issue 51

COVER STORY
Biochemistry: Proteins Containing Unnatural Amino Acids
Two new options for making designer proteins made news in 2001—both from labs at Scripps Research Institute.
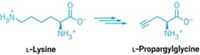
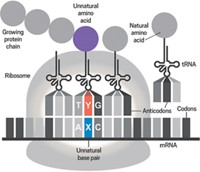
In one advance, Paul R. Schimmel coaxed bacteria with a mistake-prone transfer RNA synthetase enzyme, which catalyzes the addition of amino acids to tRNAs, to place unnatural amino acids into proteins. Meanwhile, Peter G. Schultz and coworkers inserted an unnatural amino acid into proteins made by Escherichia coli by borrowing a synthetase and a tRNA from an archaea microbe. Unnatural amino acids are protein building blocks that are not among the 22 amino acids in nature’s catalog.
Since 2001, Schimmel and Schultz have taken different directions with unnatural amino acids. Schimmel’s team has focused on the consequences of mistakes in protein production. They’ve learned that even small amounts of incorrectly made proteins in mice lead to neurodegeneration. That finding could have implications for human disease, Schimmel says.
Schultz has stayed focused on methodology. For instance, in 2001, it wasn’t clear how generalizable his technique would be to diverse unnatural amino acids. That’s not the case today, Schultz says. Researchers have incorporated more than 70 unnatural amino acids with versions of his 2001 technique or related techniques that use different synthetase/tRNA pairs.
One of those researchers is MIT’s JoAnne Stubbe, who used unnatural amino acids to prove that an electron hops over a long distance in ribonucleotide reductase, a crucial DNA repair enzyme. “There’s no way we could’ve done our studies without this technology,” Stubbe says.
Another is Jason W. Chin, a former student in the Schultz group who is now at the MRC Laboratory of Molecular Biology, in England. This year, Chin was the first to incorporate unnatural amino acids into a live animal, the worm Caenorhabditis elegans.
And at Ambrx, a biotech company Schultz cofounded, researchers have developed a long-acting version of human growth hormone as well as antibody-drug conjugates, each featuring an unnatural amino acid. The experimental hormone has completed Phase II clinical trials. A handful of other companies—including Allozyne, a firm based on science from Caltech’s David A. Tirrell—are developing unnatural amino acid therapeutics.
The scuttlebutt about unnatural amino acid incorporation has been that yields of protein are low. But Ambrx’ scientists have overcome that, Schultz says. “It’s basically being done at swimming-pool level—50,000 L.” The yields rival commercial fermentation processes, he says.
Stubbe compares the current state of unnatural amino acid technology to site-directed mutagenesis. “It took 30 years before you could take a scientist off the street and just have them make a mutant” for protein engineering, she says. But as more people are trying out unnatural amino acid technology, more kinks are getting worked out, she notes.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter