Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Diagnosing Ancient Disease
Molecules in the remains of ancient humans reveal the pathogens that plagued them
by Sarah Everts
May 20, 2013
| A version of this story appeared in
Volume 91, Issue 20
\
\
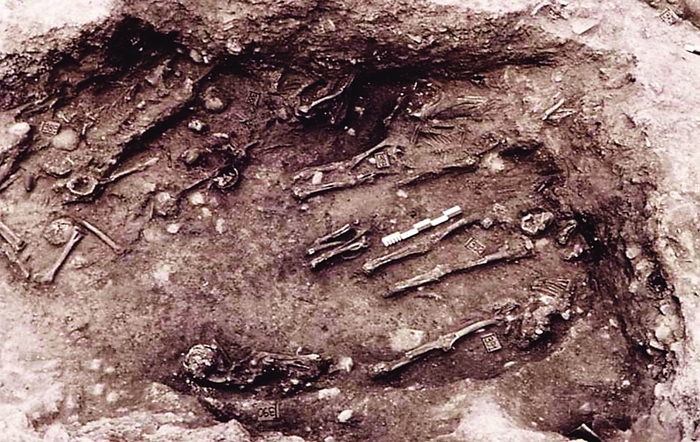
The dental remains of victims of the 430 B.C. Plague of Athens found in a mass grave revealed the pandemic was caused by typhoid fever.
In 1994, construction workers in Athens came across a mass grave containing around 150 people lying haphazardly on top of each other as if they had been buried in haste.
By dating pieces of pottery found alongside the human remains, archaeologists pinpointed the deaths to approximately 430 B.C., suggesting that the people buried in the grave were probably victims of the Plague of Athens, a pandemic of great historical significance for Western culture.
A mysterious pathogen hit Athens while it was under a Spartan siege during the Peloponnesian War that took place between the competing city-states. Nearly a third of Athens’ inhabitants and many of the soldiers trying to defend the city died in the pandemic. Many scholars point to the Plague of Athens as a contributor to the beginning of the end of the Golden Age of Greece, said Manolis J. Papagrigorakis, a Greek orthodontist who was part of a research team that studied the grave skeletons to determine the pathogen responsible for the epidemic. He described the project last month in Berlin, at a standing-room-only session of the European Congress of Clinical Microbiology & Infectious Diseases.
Until the discovery of the mass grave, the best clues about the nature of the deadly pathogen came from the Greek historian Thucydides, Papagrigorakis said. Thucydides’ gruesome account describes patients with inflamed tongues, fetid breath, skin pustules, violent spasms, and eventually, for those who did survive, the loss of fingers, eyes, and memory. The historical account helped researchers narrow down a list of possible infectious agents: Plague, typhoid, typhus, smallpox, and measles were all contenders. But to conclusively identify the Plague of Athens pathogen, researchers needed to analyze the remains of victims. So Papagrigorakis’ team analyzed the dental pulp from several individuals from the mass grave and found, in the preserved dental tissue, the DNA of Salmonella enterica serovar Typhi, the causative agent of typhoid fever.
Making infectious disease diagnoses centuries, and sometimes millennia, after the death of a patient is called paleomicrobiology, and it’s a field that’s been growing over the past two decades, said Helen Donoghue, a paleomicrobiologist from University College London, who also spoke at the conference. Before molecular techniques were up to snuff, researchers had to rely primarily on historical accounts and physical signs of infectious disease to make diagnoses. For example, long-term tuberculosis victims can have characteristic lesions on their bones and collapsed vertebrae symptomatic of tuberculosis that are still visible in 1,000-year-old skeletons, she said.
Then in the 1990s, analytical technology advanced to the point where researchers were able to analyze the DNA and other molecular residues left in or on long-dead individuals. That’s when paleomicrobiologists took things a step further and began searching for traces of pathogen DNA, proteins, and lipids in ancient remains such as skeletons and mummies.
Paleomicrobiology can answer important historical questions, Donoghue said, such as what pathogen was responsible for devastating infectious disease outbreaks in history or which infectious pathogens existed in the Americas before the arrival of European colonists.
It can also help clarify the evolution and spread of pathogens through time and space, such as whether humans first acquired tuberculosis from infected cows after animal domestication—a theory long believed to be true but recently disproven by researchers in the field.

The field adopted molecular techniques beginning in 1993, Donoghue said, when University College London researchers Mark Spigelman and Eshetu Lemma proposed that the polymerase chain reaction—which was being used at the time to amplify the human DNA found in skeletons—might also be used to detect Mycobacterium tuberculosis, the causative agent of tuberculosis, in the same remains (Int. J. Osteoarchaeol. 1993, DOI: 10.1002/oa.1390030211). After doing proof-of-principle tests on a dozen bone samples from Borneo to Britain and across the time span of millennia, the team reported that the technique could have “wide application” in helping to diagnose diseases of antiquity.
A year later, an American team reported finding M. tuberculosis in a lung sample taken from a 1,000-year-old Peruvian mummy. This research provided the first molecular evidence that tuberculosis existed in the Americas prior to the arrival of European colonists, and it helped to resolve what had been a long-standing debate about the origin of tuberculosis in the New World (Proc. Natl. Acad. Sci. USA1994,91, 2091).
Since then, researchers have identified many more infectious disease agents from a wide variety of samples. For example, scientists have shown that the 5,300-year-old Tyrolean iceman discovered in the Alps in 1991 had Lyme disease—although it was an arrow to the back that finally killed him (Nat. Commun. 2012, DOI:10.1038/ncomms1701). Other research, such as a study by Didier Raoult and his colleagues at the University of the Mediterranean, in Marseille, France, has shown that the Black Death, a pandemic that killed more than 75 million people in Europe between 1348 and 1350, was likely a coinfection of Yersinia pestis, the causative agent of the famous bubonic plague, and Bartonella quintana, which causes trench fever.
In 1998, when Raoult and coworkers found Y. pestis in the remains of Black Death victims buried in a mass grave in France, some researchers didn’t believe their conclusion that this was the cause of the plague, preferring to stick to theories that the infectious agent had been viral or a different bacterial pathogen (Proc. Natl. Acad. Sci. USA 1998, DOI: 10.1073/pnas.95.21.12637).

One reason researchers were skeptical was because plague from Y. pestis was long thought to be spread by fleas on rats. Black Death had traveled too quickly across Europe for the rat-flea infection model to be valid, especially because it had quickly traversed rural areas that did not have the rat population to account for such rapid infection rates. Furthermore, Black Death had continued to spread in winter, when rodent fleas die off, Raoult said.
Discovering that people in the French mass grave were coinfected with trench fever’s B.quintana helped Raoult’s team resolve the debate. B.quintana is spread by human lice, and in the 14th century “people were just covered in lice,” Raoult said.
So if the people in the mass grave had one infectious disease spread by human lice, maybe the pandemic’s Y. pestis was a strain that could also live in human lice instead of rat lice. That suggested Black Death had been spread by travel of humans with body lice, rather than by rats with body lice. This year, when human body lice in the Congo were found to contain a strain of Y. pestis, more people in the field accepted the theory that Y. pestis was involved in Black Death, Raoult said.
You might think that extracting the causative agents of deadly pandemics from ancient remains would make the scientists who do this research afraid of getting infected. “Of course we take the necessary precautions” to stay safe in the lab, Raoult said, but in reality paleomicrobiology researchers are often more concerned about accidentally contaminating their ancient samples with DNA from modern infectious disease agents.
That’s why the field has established a potpourri of measures to prevent contamination. When sampling from a skeleton, researchers typically extract the dental pulp, which can harbor infectious agents but is also protected from contamination over centuries or millennia by the enamel around it. Amplification of bacterial DNA found in the dental pulp is often split between multiple labs so that results can be verified.
Such contamination concerns as well as the growing sensitivity of analytical technologies have inspired researchers to look for other molecular markers for pathogenic infection in ancient remains. For example, David E. Minnikin and his colleagues at the University of Birmingham, in England, have pioneered the detection of tuberculosis pathogen lipids from skeletal remains.
Using mass spectrometry and gas chromatography, they’ve measured mycocerosates and mycolipenates in the remains of a 9,000-year-old Neolithic woman and child who suffered from tuberculosis—the oldest known cases of the disease. These fatty acids are specific to Mycobacterium, properties that are key to their use as ancient biomarkers, Donoghue said. Although proteins are often longer lasting than lipids or DNA, “it’s hard to find proteins that are specific to particular bacteria,” she added.
Because it is so difficult to find pathogen-specific proteins in ancient samples, Angelique Corthals at SUNY’s Stony Brook School of Medicine has found a way to discover the presence of pathogens indirectly. In particular, Corthals looks for characteristic human immune system proteins produced as the body tries to defend itself from a particular infectious agent.

For example, last year she reported her analysis of several 500-year-old Incan mummies that had been found in a frozen grave at nearly 7,000 feet in the Andes Mountains. Two of the mummies were so well preserved that Corthals sampled phlegm, blood, and other bodily fluids found in and near the mummies’ mouths and noses and then conducted a proteomic and genetic analysis of the fluids (PLoS One 2012, DOI: 10.1371/journal.pone.0041244).
In the case of a 14-year-old female mummy called the Maiden, samples of her bodily fluids contained DNA from a Mycobacterium species, as well as immune system proteins commonly found in chronic lung disease. In another mummy of a young boy called El Niño, the fluid in his mouth contained proteins typically produced during high-altitude pulmonary edema, a deadly condition when lungs fill with fluid.
As researchers get better at analyzing ancient human remains to find molecular proof of the sicknesses that plagued and sometimes killed them, many outstanding historical questions about past diseases and pandemics may be answered. But this research is helping contemporary humans as well. Corthals said some of the methodologies developed to study the Peruvian mummies could be adopted for forensic bodily fluid analysis. And insights into the origins and evolution of tuberculosis and other infectious pathogens may help researchers control their spread and virulence in the future.





Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter