Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
NMR Method Detects Glycosylations On Intact Proteins
Posttranslational Modifications: 2-D spectra yield fingerprints that identify stereochemistry and linkages of protein glycosylations
by Celia Henry Arnaud
May 11, 2015
| A version of this story appeared in
Volume 93, Issue 19
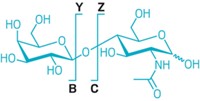
Glycosylation is the most abundant type of protein posttranslational modification. However, there’s no direct method for characterizing the chemical changes. Mass spectrometry methods, which require enzymatic digestion of the protein, can reveal the location and composition of a glycosylation, but they can’t determine the exact stereochemistry or glycosidic linkage. Mario Schubert, Michal J. Walczak, and coworkers at ETH Zurich have reported a simple and direct nuclear magnetic resonance spectroscopy method for detecting and characterizing intact glycosylated proteins (Angew. Chem. Int. Ed. 2015, DOI: 10.1002/anie.201502093). For each protein, the researchers collect a pair of two-dimensional NMR spectra—1H-1H and 1H-13C. The distinctive patterns reveal the saccharide types and linkages in each glycosylation. The analysis takes place under denaturing conditions, so the chemical shifts for all the amino acids reflect random coil structures. Any deviations from the random coil pattern indicate modifications. The team used the method to analyze glycosylated proteins from bacteria, fungi, plants, and animals. The approach works well for glycans containing up to about 20 saccharides. Because the new method can’t be used to assign a modification’s location on the protein sequence, it will be complementary to mass spec techniques.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter