Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Antibiotics
Bacterial genomes yield antibiotic candidate to replace old drug
Naturally occurring analogs to antibiotics may provide new drugs that are not plagued by resistance
by Alla Katsnelson, special to C&EN
January 13, 2022
| A version of this story appeared in
Volume 100, Issue 2

By mining bacterial genomes for metabolites distantly related to an antibiotic drug called colistin, which is now threatened by resistance in the clinic, researchers have identified and synthesized a compound that could one day replace it (Nature 2022, 10.1038/s41586-021-04264-x). Their strategy may allow researchers to identify additional antibiotic candidates, the authors say.
Many clinically important antibiotics, including colistin, are produced by or inspired by metabolites that bacteria make to kill off competing strains. Target bacteria can evolve resistance to these metabolites, and their would-be assassins in turn evolve to produce twists on those compounds to outwit that resistance.
It’s those latter compounds—naturally occurring analogs of known drugs—that Sean Brady, a biochemist at the Rockefeller University, set out to find. “Bacteria have been fighting for a long time, likely against the same resistance mechanisms,” Brady says. “Why not return to nature and see if you can find a version of that antibiotic that has gotten around it?”
The researchers searched bacterial genomes for sequences distantly related to the one that encodes colistin. These sequences are likely to produce distinct analogs to the molecule. “The most distant structure is going to be the one that has fought off the most resistance,” he says.
After identifying a candidate sequence, Brady’s group applied a computational method it had developed previously that predicts a natural product’s chemical structure from its genetic sequence. The approach allowed the researchers to home in on a compound they called macolacin. They synthesized it and close relatives with different substituent groups before landing on a biphenyl-containing analog. Biphenyl macolacin showed antibacterial properties in lab tests, and in mice it fought off infection by two colistin-resistant strains of Acinetobacter baumannii at least as well as colistin does in the absence of resistance.
“It’s a nice combination of inspiration by biology and then optimization by chemistry,” says Gerard Wright, a biochemist studying antibiotics at McMaster University. He says that though macolacin may turn out to be too toxic for clinical use, the approach can be applied to other naturally inspired antibiotics.

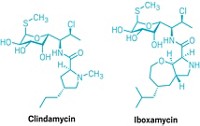
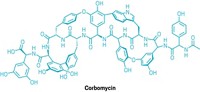
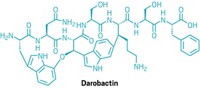

Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter