Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Computational Chemistry
Machine learning could offer chemists fast, accurate calculations
Updated neural network system predicts molecular energies and forces faster than currently used methods
by Sam Lemonick
August 22, 2018
| A version of this story appeared in
Volume 96, Issue 34
Existing computers can calculate the exact properties of only the smallest molecules thanks to the mathematical complexity of quantum mechanics. So chemists have invented methods, including force fields, density functional theory (DFT), and the coupled cluster single double triple technique (CCSD(T)), to approximate values like molecular energies and forces. Users can get quick answers or accurate ones with these methods. For example, CCSD(T) is accurate but slow, compared with force fields.
Some researchers think machine learning could offer a better way. At the American Chemical Society national meeting in Boston on Tuesday, Adrian Roitberg of the University of Florida described a method that can achieve the accuracy of CCSD(T) in the computational time of force fields. He, along with Florida colleage Justin S. Smith and Olexandr Isayev of the University of North Carolina, Chapel Hill, call it Accurate NeurAl networK engINe for Molecular Energies (ANAKIN-ME).
During a session in the Division of Computers in Chemistry, Roitberg said the third version of the method, which the team calls ANI-1ccx, can predict the forces and energy of a molecule with only the positions of its atoms and their atomic number. The algorithm treats each element separately, then produces a summed prediction of the forces and energy in the molecule as a whole.
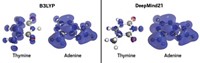
The system’s predictions were within about 0.5 kcal/mol of CCSD(T) when predicting internal rotation energies in drug-like fragments (examples shown), putting it within the top five computational methods in terms of accuracy. Roitberg said that the method obtained those results in about 2 microseconds compared with 24 hours for CCSD(T). Currently, the publicly-available version of ANI-1ccx can analyze molecules containing only hydrogen, carbon, nitrogen, and oxygen, but Roitberg said his group has added the ability to make predictions about sulfur, chlorine, and fluorine in an unreleased version of the algorithm.
To create ANI-1ccx, the researchers started with a previous version of the algorithm, which was trained on DFT predictions, then began training it on a CCSD(T) dataset. They kept any parts of the algorithm that contributed to making CCSD(T)-level predictions and replaced the others. Roitberg said that process allowed them to train ANI-1ccx on just 500,000 data points, compared with 5 million for the previous version.
If ANI-1ccx can continue to make relatively fast, accurate predictions, “there is a good chance it will become more useful than DFT for certain problems,” says Donald G. Truhlar of the University of Minnesota, who has worked on expanding the capabilities of DFT. He credits Roitberg for using a broad training set to avoid a common pitfall of machine learning algorithms, which often struggle when faced with molecules that look very different from those used in training. But he points out that ANI-1ccx may struggle with some situations that are difficult for all computational methods, like open-shell systems or excited states.
Roitberg said the group’s longterm goal is to extend ANI-1ccx to other systems, possibly starting with reactions or organometallic catalysis.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter