Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Medicinal Chemistry
Bismuth binds peptides to make bioactive bicycles
Metal-thiol complexes offer simpler way to screen bicyclic peptides
by Mark Peplow, special to C&EN
December 7, 2021

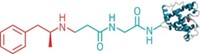
Bicyclic peptides have spurred some multimillion-dollar deals in recent years, as drug developers begin to exploit the peptides’ power to target and bind proteins.
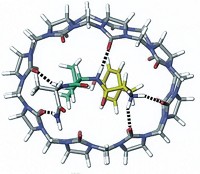
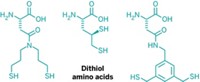
Now, researchers led by Christoph Nitsche at Australian National University have invented a new way to bend linear peptides into a bicyclic form by using a bismuth ion to tether thiol groups along a peptide’s backbone (Angew. Chem. Int. Ed. 2021, DOI: 10.1002/anie.202113857). The method could simplify efforts to screen bicyclic peptides’ interactions with proteins, and thanks to bismuth’s low toxicity, the complexes may also prove to be useful therapeutic agents.
Bicyclic peptides contain two loops in their structures. Thanks to this constrained shape, they behave like antibodies in miniature, tightly attaching to specific target proteins. These small peptides can pass through tissue and cell membranes more easily than antibodies, and their shape helps them to resist degradation in the body better than linear peptides. Several bicyclic peptides are already in clinical trials.
Researchers typically make bicyclic peptides by reacting a linear peptide with a small organic molecule that acts as a scaffold, forming covalent bonds with three amino acid residues to curl the peptide into two loops. However, the scaffold molecules sometimes form unwanted bonds within or between peptides, and they also allow some flexibility in the peptide’s structure that can reduce its targeting ability.
Nitsche’s team instead uses a bismuth(III) ion as a much smaller scaffold. The ion exclusively binds to thiols, pulling the peptide into tightly constrained loops that should be more rigid than previous bicyclic peptides. “If you rigidify it in the perfect conformation for binding,” Nitsche says, the resulting structure will have a high affinity for its biological target.
The bismuth bicycles were stable for weeks in phosphate-buffered saline, and up to 19 times more resistant to breakdown by the digestive enzyme trypsin than their linear forms.
As a proof of principle, Nitsche’s team screened 18 different bismuth bicycles against key proteases that are essential for the replication of Zika and West Nile viruses. One of the bicyclic peptides (shown) inhibited the Zika protease at nanomolar concentrations, similar to the best inhibitors reported. The same compound also inhibited the West Nile protease, and was 130 times more effective than its linear equivalent.
Using a Bi(III) ion offers a simpler route to bicyclic peptides, compared with those involving traditional scaffold linkers, says Kate Jolliffe at the University of Sydney, who studies bicyclic peptides but was not involved in the research. “In this case, they can just make the linear peptide and throw some bismuth in, and actually get a really quick comparison of their activities,” says Jolliffe. “That’s the beauty of the technique.”
“It’s such a simple way of doing it,” agrees Dehua Pei at the Ohio State University, who also works on bicyclic peptides. It’s probably also more generalizable than small-molecule scaffolding methods, he adds.
Nitsche suggests that his bicyclic peptides could also act as a vehicle for radioactive bismuth-213, an alpha particle emitter that is being tested as a targeted cancer therapy.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter