Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Protein Structure Wed to Dynamics
Technique determines structure and motions of native proteins simultaneously
by Stu Borman
January 17, 2005
| A version of this story appeared in
Volume 83, Issue 3

BIOCHEMISTRY
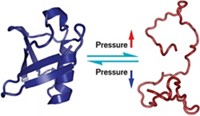
A new method for simultaneously determining the structure of a protein and the mobility and range of motion of its backbone and side chains has been developed by a British research team.
Techniques for studying protein structure and dynamics are already well developed but are generally carried out independently. The new method, dynamic ensemble refinement (DER), marries these techniques much more successfully than has been achieved in previous attempts to do so. DER could aid drug design by making it feasible to study the way small molecules interact with an ensemble of protein conformations that exist in solution—rather than with just a static, averaged structure.
DER combines molecular dynamics and two nuclear magnetic resonance spectrometry (NMR) techniques. Molecular dynamics is a theoretical technique that predicts molecular motions in proteins. NMR order parameters are an experimental measure of the possible range of such motions–that is, the extent to which side chains and other structures flop around in native (folded) proteins. And NMR nuclear Overhauser effects (NOEs) define the average conformations of native proteins. The dynamics and structure of native proteins are usually determined separately, however, making it difficult to accurately assess the entire distribution of conformations that folded proteins adopt in solution.
Research fellow Michele Vendruscolo, professor of chemical and structural biology Christopher M. Dobson, and coworkers at the University of Cambridge have now resolved this dilemma by developing DER, in which information on a native protein’s dynamics and structure are combined synergistically to map the protein's full range of conformational motion [Nature, 433, 128 (2005)].
The technique uses experimental NMR order-parameter and NOE data to rein in the molecular dynamics simulations and constrain them to reality. "It requires that an ensemble of protein conformations generated by molecular dynamics simulations is compatible with structural information, such as NMR NOEs, and also with dynamical data, such as NMR order parameters," Dobson says. The result is an ensemble of protein conformations that represents the structural and dynamic variability of the native protein.
DER could be extended to incorporate other types of experimental measurements, including NMR chemical shifts and residual dipolar couplings and X-ray diffraction data, the researchers note. The NMR order parameters used in the Nature study restricted the results to molecular motions occurring on a picosecond-to-nanosecond timescale, but using other types of measurements could enable DER to define protein motions occurring on longer timescales as well.
Chemistry professor J. Andrew McCammon of the University of California, San Diego, comments that the work"certainly represents a significant advance in structural biology. It confirms that the relatively large amplitude motions seen in molecular dynamics simulations of proteins are correct, while providing a way to improve such simulations to some degree by using experimental data to reduce errors."
NMR spectroscopist Ad Bax of the National Institute of Diabetes & Digestive & Kidney Diseases says the study "appears to represent a very significant breakthrough in that it finally presents a method that gives a quantitative meaning to the distribution of the set of NMR structures&" populated by a native protein. "This has been some sort of grail pursued by many in the past."
Indeed, University of Toronto chemistry professor Lewis E. Kay says his group and others have been trying for some time "to build a bridge between NMR experiment and site-specific contributions to thermodynamics,"but the path hasn't been clear. DER" takes a substantial step forward" toward creating that link, Kay says," and that's the real beauty of the approach."
"This is a pivotal piece of work, opening up new opportunities and insights for NMR, dynamics, protein structure, and folding,"adds biochemistry professor Jane S. Richardson of Duke University.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter