Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein Control And Modification
At 20th Protein Society symposium, researchers show how to poke, perturb, and redesign proteins
by Stu Borman
September 18, 2006
| A version of this story appeared in
Volume 84, Issue 38
This was a celebratory yearfor the Protein Society, an organization of protein scientists that's headquartered in Bethesda, Md., but has members and meetings all over the world. The year was unique in that it marked the 20th anniversary of the society's annual symposium, which alternates between Boston and San Diego and was held in the latter location last month.
What was not unique this year was the extraordinarily broad range of protein research presented at the meeting, because that kind of breadth is typical of Protein Society symposia. Topics of meeting sessions ranged from synthesis of proteins with precise modifications, optimizing protein drugs, and one-molecule-at-a-time protein sensing to designing proteins from the bottom up, analyzing the dynamics of protein function, and characterizing protein aggregates.
Dramatic advances are occurring in these and other areas of protein science. In the synthetic realm, for example, "there have truly been enormous strides made in the area of bringing tools of organic chemistry to proteins," said protein chemist Tom W. Muir of Rockefeller University, in New York City, who chaired a session on the exquisitely controlled synthesis of proteins. Two key goals of synthesizing proteins with precise modifications are to study how they work and to develop tailored versions with new functions.
Muir noted that when he entered the field of synthetic protein chemistry in the early 1990s, using organic reactions to customize proteins "was almost a pipe dream." But these techniques are now beginning to see more routine use by researchers who are not particularly knowledgeable about protein synthesis but nevertheless want to use it to study their own favorite systems.
Muir and coworkers carry out their own studies by introducing targeted changes into peptide fragments and then combining the fragments into novel proteins. They recently used such an approach to make proteins that contain nonnatural amino acids and probes, which permitted them to track movements of the proteins between different parts of living cells. "We used a photo-triggering system to follow the kinetics of movement of two different forms of a signaling protein simultaneously, one phosphorylated and one not, and showed that one goes faster than another," Muir said. "That's sort of state of the art in terms of what you can do to a protein with synthesis."
Another way to leverage organic chemistry to create bespoke proteins was described by assistant professor of chemistry Matthew B. Francis of the University of California, Berkeley. He and coworkers recently developed a new reaction that introduces a single reactive ketone or aldehyde at the N-terminus of a protein when the protein is mixed with pyridoxal phosphate (Angew. Chem. Int. Ed. 2006, 45, 5307). "It's a really mild reaction, it's easy to carry out, and the reactive group can be further derivatized using aldehyde- or ketone-specific reactions, such as oxime formation," Francis said. The researchers also recently developed a palladium-catalyzed allylic alkylation that attaches long lipid tails to proteins, a process that can be used to customize the solubility of enzymes, antibodies, viral capsids, and other proteins (J. Am. Chem. Soc. 2006, 128, 1080).
Modifying proteins with exquisite control can also be accomplished by coaxing cells into incorporating nonnatural amino acids into their proteins. "For 3 billion or 4 billion years, life on this planet has been constrained by a 20-amino-acid code," said Peter G. Schultz, director of the Genomics Institute of the Novartis Research Foundation, La Jolla, Calif. "I think we can safely say that that restriction is gone. Now it's up to the chemist's imagination, and one can think about fashioning proteins and maybe whole organisms that are not limited by the existing code."
In work carried out at Scripps Research Institute, where Schultz is a professor of chemistry, he and his coworkers developed a mutagenesis method in which a transfer RNA (tRNA) and tRNA synthetase are genetically engineered to bring a nonnatural amino acid into a protein at a single site. Schultz noted that the group has already used this type of approach to expand the genetic code by over 35 nonnatural amino acids. "I don't think there are a whole lot of constraints left right now" in the types of customized amino acids that can be incorporated into proteins, he said. Ambrx, a La Jolla-based company Schultz cofounded, is further developing the technology and plans to use it commercially.
At California Institute of Technology, professor of chemistry and chemical engineering David A. Tirrell and coworkers also specialize in nonnatural amino acid modifications. "Until a couple of years ago, the field was largely a curiosity," Tirrell said. "But Pete's lab, my group, and others have now shown a broad enough range of amino acids incorporated in several different ways that the feasibility is thoroughly established, and the need to establish feasibility has changed to a focus on utility."
Tirrell and coworkers specialize in a technique to incorporate nonnatural amino acids in which they deplete the supply of a natural amino acid in a cell and then feed it an amino acid analog. The cell incorporates the analog into newly synthesized proteins either at a single site or at many sites at which the natural amino acid would have appeared. Allozyne, a company Tirrell cofounded in Seattle, is trying to develop and exploit this approach commercially to develop medicines. "If either Ambrx or Allozyne succeeds in commercialization of protein therapeutics, people will have to take nonnatural amino acid methods more seriously," Tirrell said.
Tirrell and coworkers recently extended this approach by labeling newly synthesized proteins in a selected time window (after neuronal stimulation, pathogen infection, heat or cold shock, drug administration, or another stimulus) in such a way that they can be subsequently isolated and identified (Proc. Natl. Acad. Sci. USA 2006, 103, 9482). In this technique, called BONCAT (bioorthogonal noncanonical amino acid tagging), nonnatural amino acids with side chains (such as azides) that can undergo a specific conjugation reaction are incorporated into some fraction of positions where a corresponding natural amino acid normally occurs in newly synthesized proteins. The side chains are conjugated with a functional group that makes the proteins easy to isolate, and they are then analyzed by mass spectrometry.
Whereas Tirrell likes to create customized proteins, others prefer to watch individual proteins spectroscopically while they're actually being created. At the symposium, professor of chemistry and chemical biology X. Sunney Xie of Harvard University discussed two techniques his group developed earlier this year for observing gene expression in living cells in real time on a one-molecule-at-a-time basis (Science 2006, 311, 1600; Nature 2006, 440, 358). A dramatic movie the group created with one of the techniques shows yellow fluorescent flashes emerging from each molecule of protein at the instant of its creation from ribosomes of living cells (bernstein.harvard.edu/images/1119623clip.mov).
Single-molecule techniques like Xie's could be particularly useful for measuring the production of scarce proteins, such as transcription factors. "We're viewing bursts of individual protein expression in real time," Xie said, "making it possible to study transcription, translation, and other processes in a quantitative way."
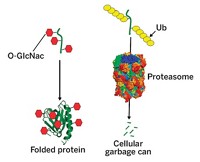
Others prefer to focus on ways proteins are modified after they are created on ribosomes. Such "posttranslational" modifications include glycosylation, acetylation, hydroxylation, and methylation. "Proteins can be extensively modified, expanding upon the structural and functional diversity encoded in the genome," said associate professor of chemistry and chemical engineering Linda C. Hsieh-Wilson of Caltech, who chaired the session. "Over 200 modifications have been identified so far, and we're just beginning to understand their biological roles."
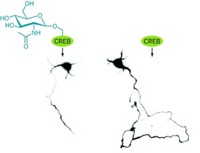
Hsieh-Wilson discussed her group's recent discovery that glycosylation of CREB (cyclic AMP-response element binding protein) inhibits the protein under hyperglycemic conditions and that this, in turn, leads to inhibition or downregulation of a gene important for survival of pancreatic cells that make insulin. This study revealed a previously unknown signaling pathway that could have implications for a better understanding of diabetes.
Some researchers just can't be satisfied with proteins as they are and instead dream of designing proteins as they might be-with novel structures and new types of functions. "Why design proteins?" asked associate professor of pharmacology Rama Ranganathan of the University of Texas Southwestern Medical Center, in Dallas, who chaired a symposium session on protein design. "We want to be able to build on, modify, improve, and change these wonderful nanoscale machines that nature has made."
Caltech professor of biology and chemistry Stephen L. Mayo and professor of biochemistry David Baker of the University of Washington, Seattle, both of whom spoke at the symposium, are among those who have had notable success in the past few years at designing novel proteins. They and others have created proteins that catalyze reactions, bind specific small-molecule substrates, act as biosensors, and bind to targets different from the native versions. Baker and coworkers recently used computational methods to design a protein fold that had never been seen in nature (C&EN, Nov. 24, 2003, page 11).
Protein design methods "are getting to the point where they could be used as tools to ask interesting questions in chemical biology," Mayo said, "but not enough of that is happening yet. There hasn't been enough focus on what can be done with these methodologies, such as designing protein therapeutics or understanding how some naturally occurring biological proteins work."
One challenge for the field is that computational design programs currently tend to be hard to use. "It's unlikely your average protein chemist is going to be able to download one of these programs and use it in the way its developers intended," Mayo said. "It's going to be a while before there are software packages out there that are user-friendly and really help you with your project."
There is but a short conceptual leap between designing proteins from scratch and improving therapeutic proteins by design. A new feature of this year's Protein Society symposium was a session on proteins as commercial drugs, highlighting "how decades of protein chemistry research have made it possible to improve commercial protein therapeutics," explained session cochair Christine E. Smith, senior director of genomics and biotechnology at Pfizer Global Research & Development, Chesterfield, Mo.

Research fellow Michael R. DeFelippis of Lilly Research Laboratories, Indianapolis, discussed how basic research on proteins has led to an important practical result: designer insulins that work better than conventional human insulin for diabetic patients. For example, Lilly's Humalog is a designed insulin with "a sequence inversion in the C-terminus of one chain that makes it faster acting," DeFelippis said. "This makes it possible to take it closer to meal time, whereas regular human insulin is supposed to be taken 30 to 60 minutes before meals. More important, the pharmacological properties of Humalog more closely match physiological insulin secretion. From a patient outcome and convenience standpoint, some of the new designer insulin formulations are a major advance in diabetes treatment."
Efforts to rationally redesign proteins would benefit from a better understanding of protein behavior, and many groups are attempting to achieve just that. For instance, researchers have been studying how the dynamics of proteins—their tendency to constantly move and change conformation—contribute to and underlie protein function.
"Proteins have evolved so they move between substates that will be productive for catalysis, signaling, or other functions," explained associate professor of biochemistry Dorothee Kern of Brandeis University, Waltham, Mass., who chaired a session on the topic. An enzyme can't function if it stays in just a single state. It has to take in substrates, catalyze reactions, and release products, and it must move to do this. A signaling protein has to go from an inactive to an active state, so a conformational change is absolutely essential there as well.
Researchers have recently begun to measure those kinds of motions, Kern said. Their findings indicate that protein motions happen on a timescale of picoseconds to seconds, depending in part on how many atoms are involved in the transitions. Local dynamics, involving small protein regions, are very fast, whereas collective dynamics, involving larger conformational changes, are slower, because different protein regions have to be in the right state for these transitions to occur.
Advertisement
Another aspect of protein behavior garnering lots of attention these days is aggregation. Proteins sometimes aggregate to form species known as amyloid fibrils. These are usually thought of as bad actors because their formation is associated with neurodegenerative conditions such as Alzheimer's disease. In a Protein Society presentation on the origins of such debilitating disorders, chemistry professor Christopher M. Dobson of the University of Cambridge, in England, noted that amyloid fibrils are not only problematic agents in biology but also fascinating nanostructures.
He outlined experiments on amyloid fibrils carried out in conjunction with colleagues in the Nanoscience Centre and the Cavendish Laboratory at the University of Cambridge. In these studies, fibrils were probed by a series of atomic force microscopy techniques, allowing some of their mechanical properties to be measured and interpreted.
"The fibrils turn out really to be extraordinary," Dobson said. "On a weight-for-weight basis, they have the same strength as steel, and their toughness is close to that of silk. These results support the view that some amyloid forms of proteins might be useful as advanced materials, rather than just giving rise to unpleasant diseases. They're infinitely 'functionizable' nanostructures with dramatic properties, and what's more, they can self-assemble in water at room temperature."
Knowledge of the mechanical properties of amyloid fibrils also has extremely important consequences for understanding how and why they are sometimes associated with pathological conditions. "The new study helps us see why amyloid deposits can be so difficult to degrade by the biological mechanisms that would normally clear this type of unwanted material from the body," Dobson said. "In addition, it provides real insight into the way amyloid can proliferate in tissue, leading to the spread of disease."
Amyloid fibrils are thus turning out to have surprising significance in disciplines ranging from nanoscience to neuroscience, Dobson noted. And in view of the extraordinary diversity of research discussed at this year's Protein Society sessions, that's something that one could say not just about amyloids but about all proteins. For proteins that are useful or just interesting, leave it up to protein scientists to poke, perturb, and redesign them in every way they can possibly imagine.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter