Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
New Way To Modify DNA
Zinc finger nickases may challenge zinc finger nucleases as genomic change agents
by Stu Borman
May 28, 2012
| A version of this story appeared in
Volume 90, Issue 22

Zinc finger nickases are the new kids on the block in the DNA-modification neighborhood. This class of DNA change agents could supplement or even supplant the more established zinc finger nucleases for some research and therapeutic applications, scientists say—but only if they can be improved. Right now that is a big “if.”
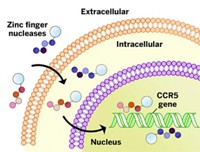
Zinc finger nucleases are synthetic proteins that are widely used to modify genes by making site-selective, double-strand breaks in genomic DNA. But they sometimes cause unintended gene changes and off-target mutations, which can interfere with experiments and lead to serious side effects when they’re used for gene therapy.
Recently developed, zinc finger nickases are variants of zinc finger nucleases that make site-selective, single-strand—instead of double-strand—breaks. So far, zinc finger nickases seem more precise and controlled in modifying DNA than zinc finger nucleases, but they are not yet as efficient as zinc finger nucleases at modifying genes. If their performance can be improved, zinc finger nickases could challenge their nuclease brethren for a range of applications.
Those applications include gene therapy, for which two zinc finger nucleases are currently in clinical trials. Sangamo BioSciences has a zinc finger nuclease that modifies CCR5—a receptor that HIV uses to infect immune cells—in Phase II clinical trials for patients with HIV/AIDS. And City of Hope is conducting a Phase I clinical trial of a zinc finger nuclease that knocks out glucocorticoid receptors in engineered T cells to treat recurrent malignant glioblastoma. Zinc finger nucleases are also widely used to generate knockouts (organisms lacking a functional gene product) and transgenic species (organisms genetically modified to express useful new properties) and to make other genetic changes for lab experiments and biotech applications.
Zinc finger nucleases cut both strands of genomic DNA at a chromosomal sequence that they are designed to target specifically. A cellular repair process called homologous recombination then takes place to make highly accurate gene modifications. The process relies on a piece of DNA—inserted into the cells at the same time as the zinc finger nuclease—as a template to copy into the genome a desired sequence change.
Sometimes, however, zinc finger nucleases are nonspecific, cutting not only the target sequence but also nontarget sequences. Another concern is that the double-strand breaks can induce nonhomologous end-joining (NHEJ), which results in random gene insertions or deletions at target sequences. In some cases, NHEJ can be desirable, but its unintended consequences can cause problems in gene modification studies and gene therapy applications.
To address the shortcomings of zinc finger nucleases, scientists began looking into nickases, which are DNA modifiers that cause single-strand breaks, or nicks.
Nickases were initially derived from homing endonucleases, another class of enzymes that create double-strand breaks. When structural biologist Barry L. Stoddard of Fred Hutchinson Cancer Research Center and coworkers, including immunology and biochemistry professor Nancy Maizels of the University of Washington, mutated a homing endonuclease three years ago, they got an enzyme that creates single-strand DNA breaks—a nickase instead of a nuclease. They found that the nickase modifies genes in human cells with about one-third the efficiency of the native enzyme (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.0810588106).
In follow-up studies, Stoddard’s and Maizels’ groups independently found that site-directed nicks do not cause the unintended mutations that accompany double-strand breaks. Thus, nicks could be safer and less toxic for gene correction. Little further study has been done on nickases based on homing endonucleases, however, because they are much more difficult to engineer for specific targets compared with zinc finger nucleases.
Now, three groups have simultaneously and independently developed zinc finger nickases, led by zinc finger experts J. Keith Joung of Massachusetts General Hospital (Nucleic Acids Res., DOI: 10.1093/nar/gks179); Michael C. Holmes of Sangamo BioSciences (Genome Res., DOI: 10.1101/gr.122879.111); and Jin-Soo Kim of Seoul National University, in South Korea (Genome Res., DOI: 10.1101/gr.138792.112).
The groups created the zinc finger nickases simply by disabling one of the two DNA-strand-cutting units found in each zinc finger nuclease. Their studies show that zinc finger nickases have little or no tendency to cut off-target sequences or cause unintended genomic changes and are as easy to customize as zinc finger nucleases.
Kim’s group found that zinc finger nickases cut fewer off-target sequences than zinc finger nucleases do. The Holmes and Kim groups reported that the random NHEJ process does not occur with nickases. For clinical applications, these characteristics mean that nickases could have more benign side-effect profiles.
Joung and coworkers did find that their nickases caused NHEJ modifications. But they studied nickase effects on synthetic genes, whereas the other two groups studied endogenous genes, where nickases would actually be used. Researchers do not yet know the mechanism of nick repair or exactly why nickases cause fewer NHEJ mutations, or perhaps none at all.
The conclusions from these three zinc finger nickase papers are similar to those of the endonuclease studies—namely, that single-strand breaks result in a reasonable amount of desired genomic changes with fewer or no unintended gene modifications, comments Daniel F. Voytas, director of the Center for Genome Engineering at the University of Minnesota. But the new studies move the endonuclease findings “to a platform that’s much easier to use” because zinc finger nickases can be more readily designed, Voytas says.
Nevertheless, zinc finger nucleases are up to about 50% effective at modifying targeted sequences, whereas zinc finger nickase efficiencies range from about 0.01 to 10%, according to the three groups. And the efficiencies of zinc finger nickases at different gene loci vary far more than those of zinc finger nucleases.
Zinc finger nuclease expert Dana Carroll says he does not believe that low efficiency “kills the nickase technology—not at all. What it means is somebody’s got to come up with a way to make zinc finger nickases much more efficient and consistent.”
Meanwhile, another potential challenger to zinc finger nucleases as targeted genomic change agents is TALENs (transcription activator-like effector nucleases), first described only two years ago by Voytas and coworkers (Genetics, DOI: 10.1534/genetics.110.120717). TALENs have the same DNA-cleaving domain as zinc finger nucleases but a different DNA-binding domain to recognize targeted DNA sequences. TALENs seem to be capable of targeting a greater proportion of specific gene-sequence targets than are zinc finger nucleases. “No one has demonstrated nicking with TALENs, but it should be trivial to do,” Carroll says. So TALEN nickases may be on the horizon too.
For now, the spotlight is on zinc finger nickases. They “have captured the field’s imagination, in terms of what their potential might be,” Holmes says. “The jury is still out a bit. There’s a good amount more work to do. But zinc finger nickases are an attractive strategy to try to control DNA repair pathway choice.”
Zinc finger nickases are “an important step forward,” says gene modification specialist Scot Wolfe of the University of Massachusetts Medical School. “It remains to be seen whether zinc finger nickases will be efficient enough for genome engineering in organisms, so it’s hard to say whether or not they’re going to completely change the landscape of what people use,” he says. “But off-target effects are clearly a concern with the current generation of zinc finger nucleases, so it’s entirely possible that nickases will supplement or supplant them for systems where one would be concerned about off-target effects.”



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter