Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Solid-State NMR: Biomolecules Viewed At Atomic Resolution
Analytical technique enables scientists to visualize membrane proteins, amyloid fibrils, components of the ribosome, and more
by Jyllian Kemsley
December 24, 2012
| A version of this story appeared in
Volume 90, Issue 52
A decade ago, researchers determined the first atomic-resolution structure of a biomolecule using solid-state NMR spectroscopy. Although the structure determination was of a mere tripeptide, says Ann McDermott, a protein NMR specialist at Columbia University, “it was a pivotal piece of work.”
Scientists in the field anticipated that solid-state NMR would be able to produce structures as detailed as those obtained by X-ray crystallography, McDermott adds. “But to see it executed that elegantly raised the bar.”

The tripeptide work was led by Robert G. Griffin, a chemistry professor at MIT, and graduate student Chad M. Rienstra, now a chemistry professor at the University of Illinois, Urbana-Champaign (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.152346599). The solid-state NMR approach incorporates magic-angle spinning, in which NMR signals are narrowed by spinning samples at a specific angle relative to the direction of the magnetic field. Since their 2002 paper, Griffin, Rienstra, and others have continued to work on the technique, with an eye toward solving the structures of proteins that are not amenable to crystallization.
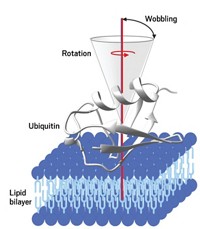
“Today the most exciting papers are targeting membrane proteins, amyloid fibrils, or large protein assemblies, such as bacterial secretion needles or elements of the ribosome,” says Lyndon Emsley, scientific director of the European Center for High Field NMR, in Lyon, France. Solid-state NMR has been particularly important for determining structures of amyloid fibrils, which are insoluble protein aggregates associated with several neurodegenerative disorders, including Alzheimer’s disease.
One key to advancing the field of solid-state NMR has been the availability of high-field magnets. Another was the development of dynamic nuclear polarization, a technique that increases the NMR signal of a sample by adding a stable radical compound. Irradiating the sample-radical combination with high-frequency microwaves transfers polarization of the radical’s electron spin to the nuclear spins of the atoms in a protein or other species of interest.
European scientists are investing more heavily in advanced NMR instruments than are scientists in the U.S., MIT’s Griffin says, largely because of funding availability. Commercial dynamic nuclear polarization and high-field instruments cost upward of $2 million, an amount that necessitates U.S. researchers gather money from multiple sources.
Griffin says he expects it will be another decade before solid-state NMR of biomolecules becomes routine. And after biomolecules, the technique’s next big hit will likely be surface science.
Two years ago, Emsley and his colleagues demonstrated the use of dynamic nuclear polarization solid-state NMR to study functionalized silica surfaces prepared as a “translucent slush” of porous silica in a radical solution. Materials scientists are interested in the approach, Emsley says, and his group is now collecting and analyzing NMR spectra of the surfaces of materials as varied as catalysts, cement, and solar-cell components.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter