Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Researchers Outline New Strategy For FRET Biosensors
Biochemistry: The technique overcomes limitations in existing FRET biosensors, possibly expanding the types of molecules that can be detected
by Laura Cassiday
August 28, 2014

A team of biochemists reports a new strategy to design biosensors that report the presence of target molecules using fluorescence resonance energy transfer (FRET). It expands the capabilities of FRET-based methods by increasing the types of proteins that can be used as the sensor (Biochemistry 2014, DOI: 10.1021/bi500758u).
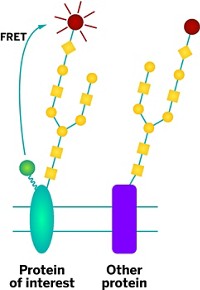
Many FRET biosensors consist of a protein with two attached fluorescent dyes. When the protein binds a target molecule, the protein changes shape and brings the dyes close together. This decrease in distance makes the energy transfer more efficient between the two dyes, which researchers can measure by monitoring the fluorescence in the cell or solution.
However, to bring the dyes close enough together for FRET to occur, proteins must undergo large conformational changes upon binding. Also, the protein must be relatively large or elongated so that, in the absence of the target, the dyes are far enough apart to prevent background FRET. Few natural proteins satisfy both of these requirements, says Stewart N. Loh, a biochemist at the State University of New York Upstate Medical University.
To expand the repertoire of molecules detectable with FRET biosensors, Loh and his colleagues decided to keep the two dyes separated completely by attaching them to different proteins. They designed the proteins so that the presence of the target brings the proteins together, allowing FRET to occur.
As a test of their strategy, the team chose fibronectin III as a scaffold for their biosensor. This antibody-like protein domain has been engineered to bind to many other proteins, one of which is the SH2 protein domain. The biochemists mutated fibronectin III’s binding site so that it no longer recognized SH2. Then, for their second protein, they used a fragment of fibronectin III with the functional SH2-binding domain. So when SH2 is present, the fragment exchanges with the mutated portion of fibronectin III, allowing the complex to bind the target and bringing the two dyes together so that FRET can occur.
The researchers tested their biosensor in buffer and found that, in the presence of SH2, the FRET signal increased more than eightfold more than the signal in the absence of the target. The team also engineered monkey kidney fibroblast cells to express the mutated fibronectin III, the fragment, and SH2. Using a fluorescence microscopy experiment, they showed that the biosensor could bind its target and produce a detectable FRET signal in cells.
Loh says that the technique should work for the more than 20 proteins that fibronectin III has been engineered to recognize. Also, he says, the strategy may work for other proteins that can be split easily into fragments.
Kevin W. Plaxco, a biochemist at the University of California, Santa Barbara, calls the strategy “a clever and promising way” to build biosensors. He notes that other groups have used similar principles to design nucleic-acid-based biosensors. However, Plaxco says, proteins offer a “richer, more complex chemistry suitable for the detection of a much wider range of targets.”




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter