Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Simple Device Identifies Cell Types By Sending Them Through An Obstacle Course
Cell Analysis: Researchers characterize the surface molecules on a cell as it travels through a microfluidic channel lined with antibodies
by Erika Gebel Berg
February 11, 2015

The forest of molecules on the surface of a cell is a kind of cellular fingerprint, unique to each cell type. To look for these molecular signatures, researchers developed a simple and inexpensive microfluidic chip that monitors cells as they travel through a channel lined with different antibodies. By measuring each cell’s transit time, the scientists can identify its surface molecules and in turn determine its type (Anal. Chem. 2015, DOI: 10.1021/ac504613b).
Flow cytometry, the standard approach for analyzing cell type, is a mainstay in the clinic and the laboratory. However, flow cytometers are large and expensive, and they require that cells be pretreated with fluorescent tags that bind to surface markers, allowing for detection. To make the identification process simpler and possibly more mobile, Lydia L. Sohn of the University of California, Berkeley, wanted to avoid hefty equipment and pretreatment. She and her team thought the solution would be to ditch fluorescence measurements in favor of detecting changes in current.
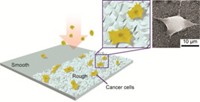
They came up with the idea while thinking about the physics of a cell moving through a microfluidic channel. Sohn’s team realized that cells would change the amount of a current running through a channel depending on the width of the channel segment. Current travels easily through cell broth, whereas cells themselves act as insulators. When cells pass through a narrow stretch, the current should drop because the cell displaces most of the liquid, effectively lowering the conductance of the channel. But in a wide segment, broth can flow around the cell, and the current is not as impeded, which appears as a spike in the current trace.
Sohn’s team used this concept to design a microfluidic channel with five narrow segments, each lined with a different antibody, built atop an electrode for generating and measuring current. The antibodies should slow the progress of any cell decorated with its antigen. Each narrow antibody zone is separated by a wider area, or node, which makes it possible to clock how long a cell lingers in each narrow segment by measuring the time between current spikes.
Sohn and colleagues then tested whether their method could identify two types of leukemia cells, NB4 and AP-1060, which express cell surface markers to different degrees. The researchers affixed antibodies along the channel that interact specifically with the antigens CD13, CD33, CD14, and CD15. They also added a segment with immunoglobulin G (IgG) as a control, as the antibody would not interact specifically with antigens on either cell type. The researchers then tested each cell line separately in their device.
Because both types of cells express CD13 and CD33, the researchers observed that most of the cells in both samples traveled slowly through the channel segments containing those antibodies, compared with their speeds through the IgG segment. Neither cell type expresses CD14, so the cells tended to race through that segment. However, the cells traveled at different rates in the CD15 region; most of the NB4 cells sauntered, whereas the AP-1060 cells sped up, relative to their respective speeds in the IgG region. The difference in speed through the CD15 segment allowed the researchers to distinguish between the cell types.
Their tests gave the percentage of cells that were positive or negative for a particular marker—results that were statistically indistinguishable from those obtained with flow cytometry, Sohn says. Each cell takes about one second to make it through the channel.
The chip design is genius, says Mark A. LaBarge of Lawrence Berkeley National Laboratory. It “allows you to track individual cells as they travel across this molecular jungle gym.” Since the approach screens cells one at a time, it’s not as prodigious as flow cytometry, which can handle huge cellular loads. But LaBarge sees the opportunity to make it a high-throughput method. Sohn is working to increase the device’s capacity with data analysis tools that can tease out transit times of multiple cells traversing the channel at once.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter