Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Microscopy
Microscopy method maps chemistry, not just chemicals
Researchers map activity of multiple enzymes simultaneously
by Celia Arnaud, special to C&EN
January 19, 2024
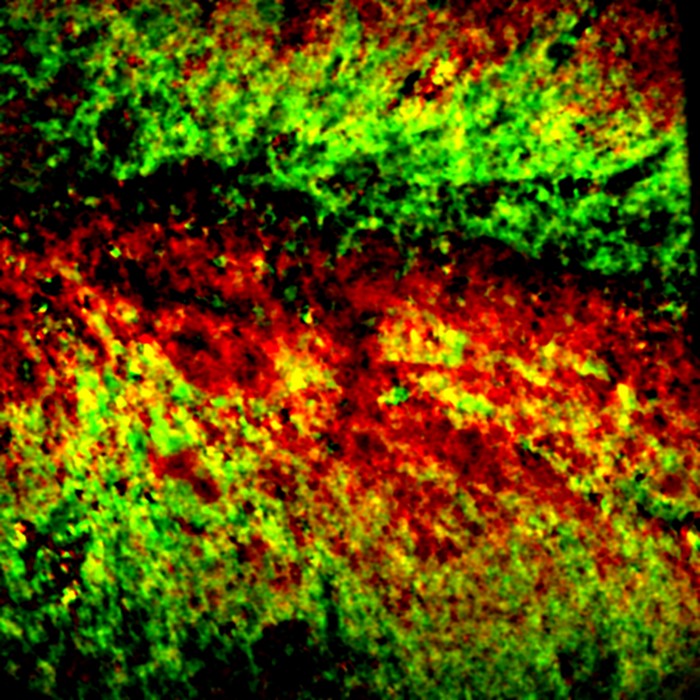
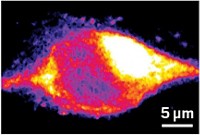
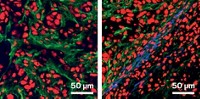
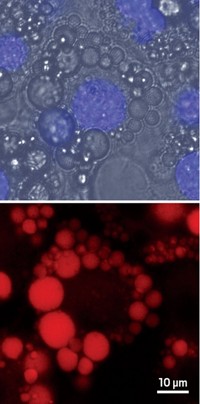
A new microscopy method enables researchers to visualize the activity of the enzymes caspase-3 and alkaline phosphatase in various biological samples (Nat. Methods 2024, DOI: 10.1038/s41592-023-02137-x) .
“We made a jump from imaging chemicals to imaging chemistry,” says Ji-Xin Cheng of Boston University, who led the research. “Chemical imaging largely maps the presence of chemicals in cells or tissues. We have developed a method to probe the substrate and the product under a high-speed, high-sensitivity vibrational microscope called MIP microscope so that chemistry can be mapped.”

MIP microscopy, which stands for mid-infrared photothermal microscopy, uses a visible beam to detect temperature effects caused by IR absorption by specific chemical bonds. Cheng and coworkers designed a new, faster MIP microscope that collects images quickly enough to do real-time imaging in living systems.
The researchers designed probes for imaging enzymatic activity. These three-part probes consist of an enzyme substrate, a nitrile reporter, and a self-assembling moiety. The nitrile reporters undergo spectral shifts between the substrate and the product, leading the researchers to dub the probes “nitrile chameleons.” The self-assembling moiety aggregates into nanofibrils that keep the product near the enzyme, which helps map the enzyme location. The signal from the product correlates with the enzyme activity.
The researchers made probes that target caspase-3 and alkaline phosphatase, both of which are involved in cancer biology. The sharp spectral peaks are far enough apart to image the substrates and products for both enzymes at the same time. “There’s cooperation between the two enzymes,” Cheng says. “This was a hypothesis in the literature for 20 years, but no one can see it. Here we provide visual evidence.”
They used MIP microscopy to measure enzyme activity in cancer cells, Caenorhabditis elegans, and mouse brain slices. The approach is as sensitive as fluorescence, Cheng says, and it avoids issues, such as photobleaching, that plague fluorescent dyes.
“Although vibrational probes have been popularly used in Raman microscopy, their use in mid-infrared imaging has been relatively rare,” Wei Min, a chemist at Columbia University who develops spectroscopic and microscopic methods for imaging biological molecules in living systems, writes in an email. “Their design of probes is novel in that they made use of aggregates in order to amplify the signal. Although the chemistry is perhaps more involved compared to simple isolated molecules, the end result appears to justify the means.”
Cheng envisions eventually being able to use the approach to image multiple enzymes involved in cellular processes such as apoptosis. “The challenge is how to design these probes so they have different peak positions, and you can monitor them inside the same cell.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter