Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Supersize Enzymes Come Into Focus
Architectures of fungal and mammalian fatty acid synthases are determined at 5-Å resolution
by Celia Henry Arnaud
March 13, 2006
| A version of this story appeared in
Volume 84, Issue 11

Fatty acid synthases, the cellular machines that produce fatty acids, are coming into sharper focus. The best structural information that has been available on these large, complex enzymes has been based on low-resolution electron micrographs. Now, a team led by structural biologist Nenad Ban at the Swiss Federal Institute of Technology, Zurich, has solved X-ray crystal structures of mammalian and fungal fatty acid synthases at 5-?? resolution (Science 2006, 311, 1258 and 1263).

Fatty acid synthases are multidomain proteins that add two carbon atoms at a time to a fatty acid chain until it has 16 or 18 carbon atoms. Besides being curious about how these enzymes do their iterative work, scientists are intrigued by them because they are potential targets for antiobesity, anticancer, and antimicrobial drugs.
The structures obtained by Ban's team, which includes postdocs Timm Maier and Marc Leibundgut and grad student Simon Jenni, "required overcoming significant technical challenges," Ban says. "Mammalian fatty acid synthase yielded only very small crystals, which, combined with the size and complexity of the molecule, made data collection almost impossible. This crystal structure was solved with crystals that only several years ago I would have considered unusable."
The fungal fatty acid synthase is a 2.6-megadalton complex, much larger than most individual enzymes, and the mammalian enzyme is a complex of two identical 270-kilodalton polypeptide chains. Both enzymes have multiple active sites.
The two proteins have markedly different architectures. The fungal synthase forms a barrel-shaped dodecamer with six each of two different polypeptides. A central wheel-and-spoke structure divides the barrel into two reaction chambers.
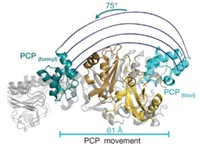
In contrast, the mammalian synthase has an X-shaped structure with two flexible "arms" and "legs" extending from its body. It also has two reaction chambers, found in the semicircular regions on either side of the body. Although the two peptides are identical, the structure is asymmetric, with substantially different size openings for the two reaction chambers.
The structures revealed some surprises. "The active sites are relatively far apart from each other, and the products of one reaction must be channeled very far in order to be used as a substrate for the next reaction," Ban says.
Craig A. Townsend, a chemistry professor at Johns Hopkins University, points out that the structures will force people to change their view of how fatty acid synthesis is carried out. Because the enzyme cycles through its reactions without releasing an intermediate, the only way the synthesis is possible is because the arms are flexible enough to bring the growing fatty acid to the different active sites, he says.

As the first reported structures of very large so-called megasynthases, these structures could be helpful in understanding other enzymes that catalyze iterative syntheses, such as the modular polyketide synthases and the nonribosomal peptide synthetases, both of which take a closely related assembly-line approach to synthesis. (In enzyme nomenclature, synthetases or synthases catalyze the synthesis of other molecules with or without the direct participation of a nucleoside triphosphate.) "Structural information about the spatial organization of these multidomain systems has been sorely lacking," Townsend says. "These papers will doubtless provide a guide both to understanding the mechanisms of these enzymes and to engineering experiments."
The current structures are a good start, according to Salih Wakil, a professor at Baylor College of Medicine who has studied fatty acid synthesis for more than 50 years, but they "are not at the resolution that everybody is dreaming of." A lot of questions remain, he says. For example, key parts of the enzymes still can't be definitively located.
Ban hopes to obtain higher resolution structures in the future. "We are determined to solve the structures at high resolution and to characterize the enzymes with respect to their interaction with reaction substrates and inhibitors," he says.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter