Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Diagnostics
Gene circuits repurpose glucose meters to detect pathogens, including SARS-CoV-2
New diagnostic tool makes glucose when infectious targets are present; an off-the-shelf glucose meter then detects it
by Celia Henry Arnaud
February 15, 2021

Researchers have co-opted glucose meters to detect infectious agents, including SARS-CoV-2, by pairing the devices with synthetic gene circuits that produce glucose in response to target analytes.
The work is “an important advance in synthetic biology toward more practical applications,” Yi Lu, a chemist at the University of Illinois, Urbana-Champaign, who has previously used glucose meters to detect other analytes and was not involved in the study, writes in an email.
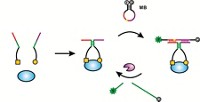
Evan Amalfitano, a graduate student in Keith Pardee’s group at the University of Toronto, and coworkers have designed gene circuits that generate glucose in response to a target analyte, usually an RNA sequence specific to a pathogen of interest. The researchers add to their system RNA that they extracted and amplified from a biological sample. The RNA binds to a “toehold switch,” an RNA hairpin loop with segments that are complementary to the target RNA. When the RNA binds, the toehold switch opens and triggers the translation of a reporter enzyme—trehalase, lactase, or phosphatase, for instance—that can convert a provided substrate to glucose. (Nat. Commun. 2021, DOI: 10.1038/s41467-020-20639-6). A glucose meter then detects the glucose produced by the enzyme. Unlike previous assays using glucose meters to detect something other than glucose, the enzymes are made during the assay rather than added as a reagent.
Such an approach can be tailored to detect most microbes by changing the target sequence in the switch. The researchers designed gene circuits to detect the Salmonella bacteria that cause typhoid and paratyphoid. They also made sensors for SARS-CoV-2 and for antibiotic resistance genes that microbes might carry, as a way to detect drug-resistant pathogens.
The researchers even made assays that simultaneously detected multiple analytes by using a mixture of gene circuits with different switches. The switches produce their products at different rates, so over a set reaction time “it is possible to see a distinction in the amount of glucose produced by which toehold switch was activated,” Amalfitano says.
At this point, the researchers can see that there’s a relationship between the amount of pathogen RNA initially present and the amount of glucose produced, but they can’t yet quantify the amount of the infectious agent present in their initial sample, he notes.
Preparing the samples for use with the circuits is a relatively involved process, which is the biggest hurdle to the system being practical, Amalfitano says. Bacterial samples require centrifugation to concentrate the cells. Breaking open the cells, amplifying the RNA, and producing the reporter enzymes all require different temperatures. To do these steps, the researchers developed a temperature-cycling incubator, which is portable but still larger than the glucose meter. Decreasing the size of the sample equipment would make the approach much more promising for other applications, Lu says.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter