Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Structural Biology
Structural biologists find path into a cancer-protecting enzyme
Insights about how HDAC10’s helix makes way for inhibitors could help researchers develop new cancer treatments
by Celia Henry Arnaud
July 22, 2020

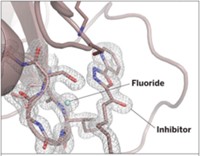
HDAC10, a member of the histone deacetylase family of enzymes, is a promising target that can help other anticancer drugs work better. The enzyme turns on processes that protect cancer cells from drugs, so researchers want to find inhibitors that can it shut down. To do that, they need to understand how the enzyme interacts with inhibitors. A new structural study led by David W. Christianson at the University of Pennsylvania helps explains the interactions that allow tubastatin A to bind and inhibit HDAC10 (ACS Chem. Biol. 2020, DOI: 10.1021/acschembio.0c00362).

For the study, Christianson and coworkers teamed up with medicinal chemist Aubry K. Miller and coworkers at the German Cancer Research Center. Other researchers at the center had done pioneering work on understanding the functions of HDAC10, discovering that HDAC10 induces autophagy—the process by which cells clear out damaged components—in response to cancer drugs. The enzyme can be inhibited by tubastatin A, but this molecule interacts with other HDACs, so Miller and other researchers want to develop more specific inhibitors that might be promising cancer drugs.
In order to make new molecules that target HDAC10, researchers like Miller want to understand how HDAC10 interacts with known inhibitors. So Christianson and Miller’s teams collaborated to solve structures of HDAC10 with tubastatin A. For this work, they used a zebrafish HDAC10 mutated at two spots to mimic the human version of the enzyme.
Researchers have been puzzled about how it is that tubastatin A can inhibit HDAC10 at all. Unlike other HDACs, the enzyme typically interacts with long, skinny polyamines—but tubastatin A is relatively bulky. Christianson and first author Corey J. Herbst-Gervasoni found a possible explanation in a key structural feature that makes HDAC10 different from other HDACs: the presence of a helix, called the P(E,A)CE motif, that constricts the active site so it works with polyamines. In the new structures, Christianson’s team found that the P(E,A)CE motif moves out of the way to make room for tubastatin A and similar inhibitors with tertiary amines.
The new work “has illuminated subtle structural features of HDAC10 that provide a blueprint for selective inhibitors of this under explored HDAC and hints for selective inhibitors more broadly,” says Stuart L. Schreiber, a chemistry professor at Harvard University who was a pioneer in studying HDACs in humans.
Most other HDACs are lysine deacetylases, but that hasn’t been the case with HDAC10. But Christianson says he now wonders whether the enzyme might be both a polyamine deacetylase and a lysine deacetylase, “at least with certain big protein substrates that can move that helix out of the way.” This possible function for the protein is worth exploring in the future, he says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter