Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Dissecting Histones from the Top down
Method combines mass spectrometry and database searching to characterize histones
by CELIA HENRY
April 5, 2004
| A version of this story appeared in
Volume 82, Issue 14

Neil L. Kelleher wants to do proteomics in a "fundamentally different way." Rather than digest proteins with enzymes before starting an analysis, Kelleher, an assistant professor of chemistry at the University of Illinois, Urbana-Champaign, takes the intact protein and lets a mass spectrometer do the work. He calls his approach, which he helped pioneer with Fred McLafferty, his graduate adviser at Cornell University, top-down mass spectrometry.
"You take the intact molecule, as presented by the biological source, and you don't degrade it with enzymes," Kelleher says. That allows you to study "all these complex molecular forms of the proteins that are expressed."
Histones are a prime example of proteins whose modifications affect their function. DNA wraps around histones, which serve as part of the packaging for the chromosomes in the cell nucleus.
"There are multiple forms of this protein, and it's really important in DNA binding and genome packaging and gene transcription," Kelleher says. The posttranslational modifications on the exposed "tails" form part of what some people call the "histone code," which may be involved in determining which genes are being transcribed.
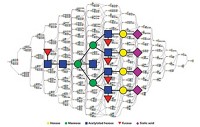
Kelleher and graduate students James J. Pesavento, Yong-Bin Kim, and Gregory K. Taylor combine a mass spectrometric approach with "shotgun annotation" to identify the modifications on human histone H4 [J. Am. Chem. Soc., 126, 3386 (2004)]. Shotgun annotation involves creating a database of various protein forms that could be generated from combinatorial consideration of different protein modifications.
"We started with the simplest histone," Kelleher says. There are five main histones in humans. H4 has seven well-known modification sites, which follow set rules for their modifications.
For the analysis, Kelleher pulled out the big gun of mass spectrometry: quadrupole Fourier transform mass spectrometry (quadrupole FTMS). FTMS uses a big superconducting magnet, making it "the Porsche of mass spectrometers," Kelleher remarks. "As if that's not enough, we stick this quadrupole on the front, and it makes the system even better. It enables us to enhance low-abundance signals." FTMS and quadrupole MS rely on different types of mass analyzers.
Once the protein is in the mass spectrometer, Kelleher and his students break the protein into smaller pieces using a method called electron capture dissociation. A lot of fragment ions are produced, Kelleher says. "We take this horrendously complicated data, and we use it to query a horrendously complicated database. The key is high mass accuracy for the fragment ions."
That database is filled with predictions of all the possible combinations of modifications. Limiting the predicted fragments "by reasonable biochemistry" resulted in a database of about 50,000 possibilities.
"We combinatorially consider all the possibilities," Kelleher says. "Now we have a technology--shotgun annotation and all this fancy mass spec--to take this very complex data and query this complex database. Out pops the right answer--automatically."
For H4, the method generated 91 fragment ions, 78 of which matched a particular prediction in the database. Kelleher believes that the other fragments didn't match because they were not within the tight mass accuracy of the database search.
Kelleher relies on the quality of the data to make sure the search algorithm correctly determines which modifications are present on a particular histone. "There's no magic wand here," he says. "If you have good data, you can have an automated characterization, but you're always limited by your data." Good data can differentiate the subtle gap between the right answer and the next-best hit.
Kelleher believes that top-down mass spectrometry makes the process of histone characterization "cleaner" than it is with other approaches. "In bottom-up methods, the peptides that are created are from a mixture of different protein forms. You garble all the different forms together in this digest and lose the connectivity of which peptides came from which protein form," he says. "The beauty of top-down MS is that you first measure these different forms and their ratios. You get a picture of the histone code at the intact level."
IN THE FUTURE, Kelleher plans to use top-down MS and shotgun annotation to track how the histone modifications vary during the cell cycle. He also hopes to pinpoint where in the genome particular histone forms are located. "You want to know if a gene is actively being transcribed in vivo, what are the histone modifications at that site in the genome? What's the code? That's where we're headed."
Alma L. Burlingame, the director of the mass spectrometry facility at the University of California, San Francisco, says, "Kelleher's method will facilitate global regio assignment of multiple modifications to individual protein species." He also believes, though, that more detailed sequencing done by another fragmentation method known as collision-induced dissociation will pinpoint modification sites in instances where electron capture does not cleave the protein in enough places.
Nevertheless, Kelleher is evangelistic about proteomics using top-down MS. He has created a website for top-down proteomics with a protein database that emphasizes posttranslational modifications using a program called ProSight PTM. His website currently has about 100 users.
"Top down is going to have a role to play in proteomics, and that role is going to do nothing but grow," he says.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter