Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Environment
Designer DNA snaps into holey crystals
May 15, 2006
| A version of this story appeared in
Volume 84, Issue 20
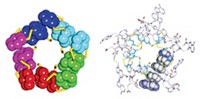
While attempting to cocrystallize DNA and protein molecules, Paul J. Paukstelis, a crystallographer with the Institute of Cellular & Molecular Biology at the University of Texas, Austin, was disappointed: Only the DNA crystallized. Then Paukstelis wondered if he could design DNA oligomers that would assemble into crystals with a regular network of pores that could host hard-to-crystallize proteins and other molecules with a regularity amenable to crystallographic analyses. By designing oligomers that exhibit both classic binding between C−G and T−A nucleotides and nonclassical pairings such as A−A and G−G, he has taken "a first step," he reports. When he expands these "assembly strands" with spacer strands, the expanded oligonucleotides self-assemble into sugar-grain-size crystals with long-range, 9-nm pores along one of the crystal's directions. Paukstelis reports that proteins below a threshold size infiltrate these pores but larger molecules do not, a trait important for the applications he has in mind (J. Am. Chem. Soc., published online May 9, dx.doi.org/10.1021/ja061332r).





Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter