Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Synthesis
Largest Chemically Made Protein
HIV protease analog assembled using kinetic ligation approach
by Stu Borman
February 22, 2007
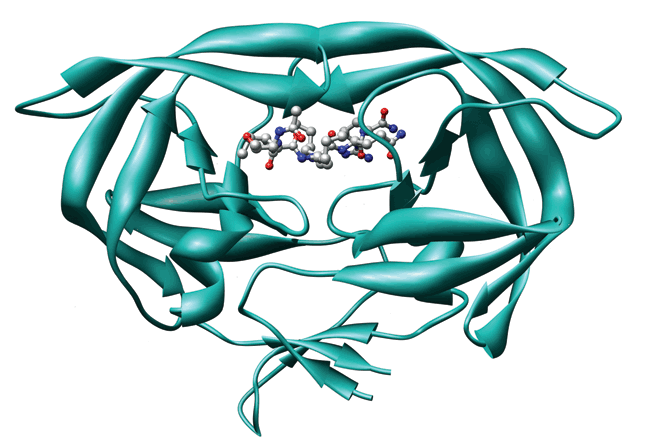
With a protein synthesis approach that they first used and reported last year for making a small model protein, researchers now have assembled large protein analogs from easily modified components.
The technique, kinetically controlled ligation (KCL), was developed by chemistry professor Stephen B. H. Kent of the University of Chicago and coworkers, who have now extended its applicability to the synthesis of a HIV protease analog (Angew. Chem. Int. Ed. 2007, 46, 1667). "To the best of our knowledge, the 203-amino-acid HIV protease covalent dimer is the protein with the largest linear polypeptide chain ever prepared by total chemical synthesis," Kent says.
The technique could make it easier to create protein analogs modified virtually at will for studies of protein function and mechanism and as drug candidates.
Kent believes the largest protein made previously by chemical synthesis was a 166-amino-acid erythropoiesis protein that his group created in 2003. That protein was made sequentially—working linearly from one end to the other—using an earlier technique, native chemical ligation (NCL). The power of KCL is that it makes it possible to synthesize proteins convergently—assembling large pieces that are then linked to one another—which is much more efficient and more practical than sequential synthesis.
Kent and graduate student Vladimir Torbeev created the 22-kD HIV protease molecule by synthesizing four peptide segments via stepwise solid-phase synthesis, using KCL to combine the segments into two large fragments, and then using NCL to combine the fragments.
The product is a single polypeptide chain that folds to form an HIV protease analog with the full enzymatic activity of the native version. The synthesis "illustrates the potential of the convergent synthetic strategy for making larger, more complex protein targets," Kent says.
"The work is a tour de force in protein chemical synthesis that showcases the state of the art in peptide ligation technology," comments Tom W. Muir, head of a protein engineering group at Rockefeller University.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter