Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Physical Chemistry
Enzyme's Active Site Sighted
Methane monooxygenase may use dicopper center to make methanol
by Stu Borman
April 28, 2010

A research team believes it has solved a long-standing controversy in the field of bioinorganic chemistry about the identity and location of metal atoms in a key methane-oxidizing metalloenzyme (Nature, DOI: 10.1038/nature08992). But the results are being challenged strongly by at least one group.
If verified, the findings—by metalloprotein specialist Amy C. Rosenzweig of Northwestern University and coworkers—could lead to the development of protein catalysts capable of oxidizing methane to methanol much more efficiently than is now possible. Methanol is a potential alternative to oil and natural gas for the production of organic chemicals and fuels.
“Vast world reserves of methane gas are underutilized as a feedstock for the production of liquid fuels and chemicals owing to the lack of economical and sustainable strategies for the selective oxidation of methane to methanol,” the researchers note in their paper. Current processes to activate the strong C–H bond in methane “require high temperatures, are costly and inefficient, and produce waste,” they add.
Metalloenzymes called methane monooxygenases (MMOs) from methanotrophic bacteria activate methane efficiently at ambient temperature and with no waste. Scientists would like to better understand them because they might point the way to an efficient, environmentally benign catalytic C–H oxidation process.
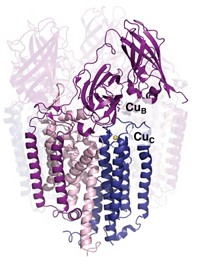
But there are two types of MMOs, and although the di-iron centers of the first type, soluble MMOs, are well-known, the metal composition and active-site location of the other type, particulate MMOs (pMMOs), have been a matter of controversy for about 20 years.
Previous studies have proposed that pMMOs have iron or copper ions in various numbers and in different active-site locations, including di-iron and tricopper centers within the enzyme’s membrane domains. Rosenzweig and coworkers now show that pMMOs have a dicopper active site located in the enzyme’s soluble domain. They removed all the metals from pMMO and showed that adding back copper, but not iron, restores most of the enzyme’s activity. They then made a protein fragment that includes the soluble dicopper-containing domain but not the membrane domains and showed that the fragment oxidizes methane.
However, enzyme biochemists Sunney I. Chan and Steve Sheng-Fa Yu of Academia Sinica, in Taipei, are questioning the findings. They note that the soluble protein fragment has far less activity than a form of natural pMMO that has 15 copper ions, including an intramembrane tricopper site, and two active sites. In their view, Rosenzweig’s reconstituted dicopper system does not by any means recapitulate the activity of that form of natural pMMO, calling into question the new study’s conclusions. In addition, Chan and Yu believe the study dismisses too quickly the possibility that iron, as well as copper, might be needed for activity of the enzyme.
Rosenzweig replies that there is “not strong evidence for an intramembrane tricopper cluster or for the binding of 12 additional copper ions” in native pMMO and that “it is unlikely that it would have two sites capable of oxidizing methane.” She believes her group used an appropriate form of native pMMO as a standard of activity. And she notes that the activity of her group’s reconstituted fragment is only modestly reduced relative to that standard—probably because the fragment is missing the native enzyme’s membrane regions, which may contribute to the enzyme’s activity. She also believes her group’s experiments rule out catalytic contributions from iron.
Others view the new study more favorably than do Chan and Yu. “The results provide direct, convincing evidence that rules out iron as being required for activity, argues against tricopper clusters as being catalytically relevant, and shows that the dicopper site is the likely site of methane oxidation,” says William B. Tolman, a specialist in metalloprotein active sites at the University of Minnesota. “The findings open the door to exciting future work on pMMO’s chemical mechanism—for example, what is the structure of the active site in its active state and how does it perform methane oxidation?” It’s likely this work will stimulate follow-up by many others who work on the enzyme and on synthetic model complexes.
And J. Martin Bollinger Jr. of Pennsylvania State University, a specialist in metalloenzyme mechanisms, says, “Everybody knew that soluble MMO, which has been very well characterized, has a di-iron center, but the issue of iron versus copper in pMMO was really up in the air.” The methodology of Rosenzweig and coworkers was so painstaking and meticulous that “I think their study is absolutely conclusive” in identifying a copper catalytic center, Bollinger says.
Whether that is the case may take more time to determine. And with huge supplies of methane awaiting oxidative liberation, it’s a question that researchers will most likely continue to pursue.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter