Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Physical Chemistry
Mediator Gives Up A Few Secrets
Structural Biochemistry: View of transcription complex could aid mechanistic studies, drug discovery
by Stu Borman
July 11, 2011
| A version of this story appeared in
Volume 89, Issue 28

In a major step toward achieving one of the key unsolved problems of structural biochemistry, the crystal structure of a large part of the yeast Mediator protein complex has been determined.
Found in eukaryotes from yeast to humans, Mediator plays a role in the expression of genes and their transcription into mRNA. The structure will help researchers learn more about how it assembles from constituent proteins and how it works at a molecular level, which has been largely unknown. The work could also aid discovery of drugs that control gene expression by targeting Mediator, just as some antibiotics target the ribosome.
Transcription factors known as activators regulate gene expression by binding to gene-promoter regions and encouraging RNA polymerase II-catalyzed transcription of the genetic code into mRNA. In 1990, a group led by Roger D. Kornberg of Stanford University School of Medicine found that an enormous protein complex they called Mediator was responsible for passing instructions from activators to RNA polymerase II.
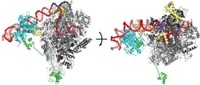
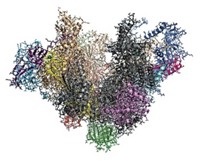
Scientists have chased Mediator’s makeup ever since, but the best structures to date have included only two or three subunits; whereas yeast Mediator has 21 and those of higher eukaryotes have more. Now, biochemist Yuichiro Takagi of Indiana University School of Medicine and coworkers have crystallized and structurally analyzed yeast Mediator’s seven-subunit “head,” a component essential to its activity (Nature, DOI: 10.1038/nature10162).
A surprising revelation is that 10 α-helices are shared by five of the head’s subunits—the first such bundle found to originate from so many proteins, the researchers note. The structure provides clues about how Mediator assembles, how it’s stabilized, and how it works.
“Mediator function is central to all or most RNA polymerase II transcriptional regulation, but its mechanisms are only just being worked out,” comments RNA polymerase II expert Joan Conaway of Stowers Institute for Medical Research, Kansas City. “This structure is a really big deal because it should enable and guide the rational design of experiments to discover possible mechanisms.”
Challenges remain: Mediator’s “middle” and “tail” have yet to be determined, the structure’s 4.3-Å resolution isn’t sufficient to resolve atoms, and parts of the head aren’t included in the model.
But the structure is a major achievement “because Takagi and coworkers had to express the subunits, assemble the head module, and then get crystals that would diffract,” says Mediator specialist Michel Werner of the Institute of Biology & Technology, in Saclay, France. “It shows how the subunits are arranged and helps in interpreting how Mediator recruits transcription factors to activate polymerase.” The work suggests it will eventually be possible to get a complete picture of the Mediator complex, he says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter