Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Pyrrolysine Synthesis Revealed
Biochemistry: Work provides a deeper understanding of amino acid evolution
by Celia Henry Arnaud
April 1, 2011
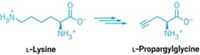
Biochemist Joseph A. Krzycki and coworkers at Ohio State University (OSU) have figured out that lysine is the only precursor required for the biosynthesis of pyrrolysine, the 22nd genetically encoded amino acid and the last one to have its biosynthetic pathway determined (Nature, DOI: 10.1038/nature09918). The work provides a deeper understanding of amino acid evolution and could lead to the creation of novel amino acid derivatives.
Pyrrolysine, which was discovered in 2002 in methane-producing Archaea, consists of a lysine chain connected to a methylated pyrroline ring. The methyl group made the pyrroline part of the molecule a biosynthetic mystery. Biochemists guessed that it could come from cellular metabolites like methionine, ornithine, glutamic acid, proline, or isoleucine.
The OSU researchers incorporated archaean genes for a methyl-transferring enzyme that has a pyrrolysine amino acid residue and for the enzymes involved in pyrrolysine synthesis into Escherichia coli and fed the bacteria lysine labeled with heavy isotopes of carbon and nitrogen. Mass spectrometric measurements of the methyltransferase biosynthesized by the engineered bacteria revealed that all the carbon and nitrogen atoms in its pyrrolysine amino acid were heavy isotopes, indicating that lysine was the sole precursor.
In the biosynthesized pyrrolysine, "we found that the lysyl came from lysine as you would expect, but the ring came from lysine, too," Krzycki says. "That was not expected."
In the proposed pathway, the enzyme PylB converts an L-lysine to (3R)-3-methyl-D-ornithine. A second enzyme, PylC, adds another L-lysine to the ornithine intermediate. A third enzyme, PylD, eliminates ammonia from the molecule, yielding (3R)-3-methyl-D-glutamyl-semialdehyde-Nε-L-lysine, which undergoes a cyclization reaction and loss of water to produce L-pyrrolysine.
They found that if they skipped the first step and added D-ornithine and lysine, PylC and PylD still worked, but the final product was desmethylpyrrolysine, in which pyrrolysine's methyl group was replaced by a hydrogen atom. The fact that the two enzymes function with something other than their normal substrate suggests that the system could be used to biosynthesize other unnatural amino acids as well, Krzycki says.
"The biosynthetic pathway proposed by the authors is reasonable and chemically feasible," Steven W. Ragsdale writes in an accompanying commentary. "The door is now open for enzymologists to study the [pyrrolysine] biosynthetic pathway in detail."
On a more fundamental level, the new pathway could help biologists understand how amino acids evolve and become incorporated in the genetic code.
"Understanding how a particular amino acid is synthesized sheds light on the evolution of the pathway involved and so on the history of that amino acid," says John F. Atkins, a geneticist at the University of Utah.
"It is exciting to learn from nature such a simple and elegant scheme for introduction of new amino acids into the genetic code," says Dieter Söll, a biochemist at Yale University. "I am convinced that scientists will imitate this concept with their own designs."



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter