Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
A Genetic On-Off Switch Enables Study Of Newly Minted Proteins
Proteomics: Unnatural amino acids tag proteins that a cell makes when its environment changes
by Olga Kuchment
June 20, 2012

A new technique allows researchers to pinpoint proteins that a cell makes when responding to other cells or its surroundings. (ACS Chem. Biol., DOI: 10.1021/cb300238w).
Erin Schuman, of the Max Planck Institute of Brain Research, in Germany, and David Tirrell, a chemist at California Institute of Technology, teamed up to develop the method to study at a molecular level how memories form. Schuman’s research group wanted to identify the proteins that a neuron creates immediately after receiving a signal from another neuron.
Unfortunately, existing techniques to identify freshly made proteins are not sensitive enough to spot the low concentrations of proteins made after neuron signaling. One common proteomic method grows cells in the presence of 13C-labeled amino acids. Pinpointing all the labeled proteins in a mixture including unlabeled ones is not always possible, Tirrell says.
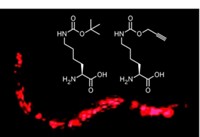
As a solution, Schuman, Tirrell, and their colleagues refined a way of incorporating unnatural amino acids into newly made proteins. The side chains of these amino acids not only distinguish the proteins that contain them from others in the cell, the researchers thought, they also could provide a handle for researchers to use to isolate the new proteins.
To trick the cell into incorporating unnatural amino acids into proteins, the researchers relied on an enzyme they had developed previously: a mutant tRNA synthetase, which attaches amino acids to tRNA molecules. They had designed the enzyme to add azidonorleucine to a tRNA that normally carries methionine (Nat. Chem. Biol., DOI: 10.1038/nchembio.200).
In the current work, the team found a way to activate this enzyme only during specific cellular events. They combined the gene for their enzyme with a genetic element called a promoter that determines when the cell transcribes a gene. The promoter they added turns on only when the cell senses an environmental change, in this case the onset of oxidative stress.
To test their method, the scientists added a DNA plasmid containing the mutant enzyme gene and the oxidative-stress promoter to Escherichia coli cells. They then exposed the cells to the herbicide paraquat, which produces free radicals, and added azidonorleucine. After 15 minutes, they broke open the cells and separated all of the cells’ proteins by gel electrophoresis. To help spot the new proteins, they added a fluorescent dye that would react only with the unnatural amino acids’ azido groups.
When the researchers added azidonorleucine to cells that they had not exposed to paraquat, they saw only a small number of tagged proteins. The amount of labeled proteins increased with how much paraquat the scientists added, indicating to them that the environmental signal triggered the cell to incorporate the unnatural amino acid into the proteins.
Tirrell says that, instead of a dye, the researchers could add molecules such as biotin to the tagged proteins. These molecules would allow the scientists to pull out the new proteins from the cell mixture.
Compared to existing methods, the new method seems to provide a much more complete picture of changes in protein synthesis inside a cell, says Samuel Gellman, a chemist at the University of Wisconsin, Madison. It could allow researchers to see all the proteins synthesized when a specific promoter turns on, he adds.
Tirrell sees a number of applications for the method. He has begun to use it to study protein synthesis during biofilm formation. Tirrell thinks it also could uncover new antibiotic targets by spotting proteins that bacteria make when infecting their host.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter