Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Spectroscopic Technique Spots Effects Of Deamidation On Proteins
Protein Structure: Spontaneous side-chain transformations in amyloid proteins cause subtle structural changes
by Melissae Fellet
July 6, 2012

Enzymes inside cells tweak and modify amino acids to change proteins’ structure and function. But some amino acids also undergo a transformation called deamidation without the help of an enzyme. Scientists want to understand how this spontaneous reaction changes protein structure, because they think it may play a role in disease. Now scientists have detected subtle structural changes, unseen by standard methods, in deamidated aggregates of a peptide linked to type-2 diabetes (J. Am. Chem. Soc., DOI: 10.1021/ja3039486).
That peptide, amylin, is a hormone that helps regulate glucose levels in the body. In patients with type-2 diabetes, amylin aggregates to form short fibrils that damage insulin-producing cells in the pancreas. Scientists have spotted deamidated versions of the peptide in the lab but they didn’t know whether the modification affects the fibril structure, says Niels Andersen, of the University of Washington, Seattle, who was not involved in the new study.
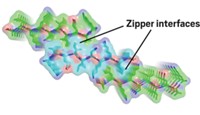
In any peptide, only two types of amino acids can undergo deamidation: glutamine and asparagine. During the reaction, their amide side chains spontaneously convert into carboxylic acids. This modification adds a negative charge to the peptide and can kink its backbone, scientists think. Most techniques used to study amyloid fibril structure, such as microscopy and circular dichroism spectroscopy, aren’t sensitive enough to pick up such a small structural change.
So Martin Zanni, of the University of Wisconsin, Madison, and his colleagues developed a new method using two-dimensional infrared spectroscopy. They synthesized four versions of the amylin peptide, each with a single carbonyl group in its backbone labeled with 13C and 18O. Using a method that included mass spectroscopy, the scientists confirmed each peptide deamidated at one or more of amylin’s one glutamine and six asparagine residues. Transmission electron microscopy showed that the peptides aggregated into fibrils.
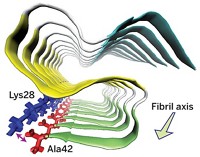
Then the scientists scanned deamidated amylin fibrils with infrared light and looked for a signal in the resulting spectrum that corresponded to the labeled carbonyl. Based on the position of this signal, the researchers could determine whether that amino acid was ensconced in a properly folded β-sheet structure or was in an unfolded state. A labeled amino acid near the center of the peptide produced a signal indicating to the researchers that the region of the peptide was folded properly. However, amino acids at the ends of the peptide appeared to be in an unfolded state.
The scientists also tracked the aggregation of amylin peptides by watching for a IR signal that indicates fibril formation. They found that deamidated amylin forms fibrils faster than the unmodified peptide does, an effect they think may be due to structural instability induced by deamidation.
Andersen applauds the level of structural detail the method reveals. But while deamidation speeds fibril formation in the lab, he points out that no one has spotted deamidated amylin in the body.
Because other amyloid-forming proteins deamidate inside the body, Zanni thinks amylin likely deamidates in the pancreas. He hopes the structural information the new technique provides could help people design drugs to prevent protein aggregation.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter