Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
New Starting Point For Flu Drug Design
Structural Biology: Different flu antibodies interact with viral protein in similar way
by Celia Henry Arnaud
February 18, 2013
| A version of this story appeared in
Volume 91, Issue 7

Influenza viruses enter cells by using spiky proteins called hemagglutinins to bind with cell-surface receptors that are tipped with sialic acid. Blocking this interaction is an obvious strategy for flu therapies, and many drug discovery efforts start with a sialic acid scaffold to block the hemagglutinin binding pocket. The problem: Sialic acid is a lousy binder.
What’s needed is a different starting point. New structures of human antibody fragments bound to H2N2 flu virus, a type of Asian flu, could point the way to new scaffolds. (The H and N stand for hemagglutinin and neuraminidase, respectively, and the numbers determine the virus subtype.)
“In the circle of influenza aficionados, the H2N2 subtype is considered to be a dormant giant,” says Wayne A. Marasco, an influenza researcher and professor of medicine at Dana-Farber Cancer Institute and Harvard Medical School, who was not involved with the current structure study. The Asian flu virus, which caused a 1957 pandemic, stopped circulating in humans in the late 1960s but continues to reside in birds and pigs. Scientists worry that it could jump from animals back to humans, sparking another pandemic.
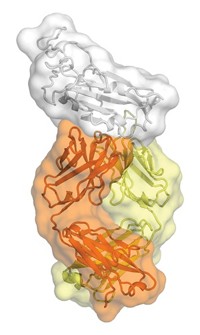
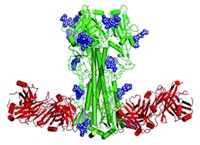
Structural biologist Ian A. Wilson of Scripps Research Institute, immunologist James E. Crowe Jr. of Vanderbilt University, and coworkers have now solved crystal structures of H2 hemagglutinin complexed with the antigen-binding fragment of three human antibodies that neutralize the 1957 Asian flu virus (Nat. Struct. Mol. Biol., DOI: 10.1038/nsmb.2500).
“Even though the antibodies approach from different directions and have completely different sequences, they’re actually finding commonality in the way they interact with the receptor-binding site,” Wilson says.
In each case, the antibody inserts an aromatic ring into a hydrophobic pocket in the receptor-binding site. The aromatic ring interacts with a tryptophan residue in the site via π-π stacking. The researchers also showed that hemagglutinin mutations that enable the virus to evade the antibodies greatly reduce the protein’s ability to bind sialic acid.
The work shows that the hemagglutinin receptor-binding site is “an important mold for drug engineers in their search for new scaffolds for small-molecule drug design,” Marasco says. In terms of vaccines, the structures expand the map of binding sequences that neutralize hemagglutinin and “can serve as a blueprint for novel antigens and as a measure to characterize and judge antibody responses to future H2N2 vaccines.”



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter