Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Enzymes Steer Toward High Substrate Concentrations
Molecular Motors: Researchers think this enzyme motion could drive self-propelled nanomotors
by Sarah Webb
January 25, 2013

Researchers imagine autonomous nanosized devices sailing through the blood stream, detecting molecular signs of disease or delivering drugs. Such devices will need nanomotors that can drive and steer them through the body. Now a team at Pennsylvania State University has shown that enzymes could act as molecular-sized engines. As enzymes catalyze reactions between molecules, the proteins propel themselves toward high concentrations of the reactants, the scientists report (J. Am. Chem. Soc., DOI: 10.1021/ja3091615).
A critical challenge for building catalytic, self-powered nanomotors is translating the energy released by chemical reactions into directed motion, says Ayusman Sen of Penn State. Recently he and his colleagues showed that enzymes diffuse faster in solutions that include their substrate, the small molecule whose reaction they catalyze. So the team wondered if an enzyme would follow an increasing concentration gradient of its substrate, just as bacteria trace a chemical trail toward a food source. If so, such gradients could steer enzyme movements.
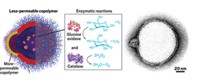
Working with Peter J. Butler, a bioengineer at Penn State, the researchers labeled two different enzymes, urease and catalase, with fluorescent dyes and watched each enzyme’s movements in a microfluidic device using fluorescence correlation spectroscopy. In the device, solutions from two input channels flow side by side in a main channel. One solution contained the labeled enzyme and the other had the enzyme’s substrate.
The scientists monitored how far the enzymes diffused from the enzyme side of the main channel across the boundary between the two incoming solutions. The distance the enzymes moved depended on the concentration of the substrate solution the researchers pumped into the device: Higher concentrations, on average, caused greater enzyme diffusion. Sen and Butler’s team didn’t observe this concentration-dependent diffusion when they inhibited catalase, suggesting that the reactions catalyzed by the protein somehow fueled the movement. Currently, the researchers don’t have an explanation for the force driving this motion.
In another experiment with the microfluidic device, the researchers paired the labeled catalase solution with one containing glucose oxidase. Glucose oxidase reacts with glucose to produce hydrogen peroxide, which is catalase’s substrate. The team observed that the catalase proteins diffused deep into the glucose oxidase side of the main channel only when they added glucose to the mix.
Henry Hess of Columbia University calls the findings intriguing and surprising. He thinks that the glucose oxidase experiment offers a possible insight into the organization of prebiotic synthetic systems. Without cellular membranes to corral the collection of catalysts needed to synthesize a complex molecule, Hess wonders if they could have self-organized, as each catalyst diffused toward the molecules producing their substrate.
Sen and his colleagues next want to see if they can use substrate concentration gradients to direct nano- or microparticles decorated with enzymes.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter