Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Physical Chemistry
The Protein ‘Periodic Table’
Structural Biology: Analysis predicts Interfaces And Arrangements Of Complexes
by Celia Henry Arnaud
December 21, 2015
| A version of this story appeared in
Volume 93, Issue 49
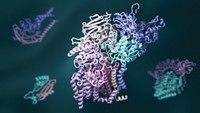
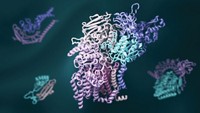
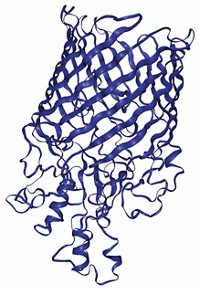
Researchers have created a “periodic table” that enables them to organize and predict a range of possible quaternary structures for protein complexes. Understanding such structures can provide insight into protein function and evolution. A team led by Sarah A. Teichmann of the European Bioinformatics Institute and the Wellcome Trust Sanger Institute constructed the table from experimental and computational data about the assembly pathways of homomeric and heteromeric complexes (Science 2015, DOI: 10.1126/science.aaa2245). The researchers identified five types of assembly steps, which they used to elucidate all possible patterns of subunit interfaces for complexes with given stoichiometries. They organized the resulting structures into a table ordered by number of subunit repeats and the number of unique subunits in a given complex. Approximately 92% of all known protein complex structures fit into the model; the other 8% may indicate structure assignment errors, the researchers note. The table could help in the bioengineering of proteins by showing what combinations of subunits and interfaces would form a complex with a desired arrangement.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter