Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Discovery of covalent ligands on the proteome-wide scale
Fragments bind to proteins previously thought undruggable
by Celia Henry Arnaud
June 20, 2016
| A version of this story appeared in
Volume 94, Issue 25
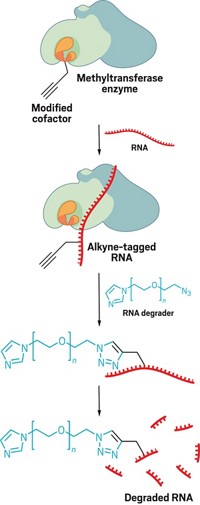
There may be an untapped reservoir of “druggable” protein targets, according to a new study. To help find potential drug targets, Benjamin F. Cravatt, Keriann M. Backus, and coworkers at Scripps Research Institute in La Jolla, Calif., have extended a method called fragment-based ligand discovery to a proteome-wide scale in native biological systems and used it to identify many proteins that weren’t previously known to bind ligands (Nature 2016, DOI: 10.1038/nature18002). The researchers exposed intact cells or cell lysates to protein-binding molecular fragments attached to electrophilic groups that covalently react with cysteine. Then they treated the proteins with an alkyne-labeled probe known to react with many cysteines in the proteome. If a fragment is already bound to a cysteine, that cysteine is described as “liganded,” and the fragment blocks the probe from binding. In 637 proteins, the researchers identified 758 liganded cysteines, which they defined as ones for which probe binding was reduced by at least 75% relative to samples exposed to dimethyl sulfoxide (DMSO) instead of the fragments. More than 85% of the proteins thus identified were previously not known to bind ligands. For example, the researchers identified ligands that target and distinguish between the inactive precursor forms of two caspases, a family of enzymes that are involved in cell death.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter