Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
A faster way to find medicines hiding in nature
Crystallography approach creates a shortcut for capturing and characterizing enzyme inhibitors from wild microbes
by Erika Gebel Berg
August 12, 2016

Pharmacies are filled with molecules inspired by nature, but finding new drugs in the wide world is expensive and time consuming. Now, researchers have developed a streamlined process for identifying drug-target enzyme inhibitors and solving their molecular structures, an approach that may speed the development of new therapies (J. Nat. Prod. 2016, DOI: 10.1021/acs.jnatprod.6b00215).
Organisms such as bacteria, plants, insects, venomous reptiles, and more produce a multitude of molecules with potential medicinal activity. To find natural products that interact with drug targets, scientists typically sift through biomass via a lengthy series of extractions and purifications. At each step, the researchers check the fractions for activity against a target. Then, with a molecule in hand, they characterize its structure using spectroscopy, X-ray diffraction, and other analytical techniques. The process is expensive and can take years, says Dieter Bromme of the University of British Columbia.
To speed up the process, his team developed an approach that uses a protein drug target to fish promising molecules out of a relatively crude natural extract early in the process. The team performs the minimum purification that allows them to use crystallography to characterize the protein complex formed when a molecule inhibits the drug target. The process reveals the structure of the natural product in a matter of weeks.
Bromme’s team tested their method using cysteine protease cathepsin K, an enzyme involved in the breakdown of bone tissue and a drug target for treating osteoporosis; inhibiting this protein could reduce bone loss. Using an assay that detects the protease’s activity, the researchers screened a library of 350 bacterial cultures isolated from lichen and soil from the rain forests of British Columbia for inhibitory activity against cathepsin K. They partially purified the crude extract from the strain with the most potent inhibition, a Streptomyces species, and then mixed the extract with cathepsin K. To see what they’d caught with their protein bait, the researchers co-crystallized the inhibitor-protein complex. The structure revealed a novel high-affinity inhibitor, lichostatinal.
For comparison, the researchers also performed the traditional extraction of cell cultures, purification of inhibitors from active fractions, and structural characterization of the resulting products, but they didn’t come up with lichostatinal. Instead, that approach yielded a known inhibitor of cathepsin K, antipain, which is likely much more abundant in the bacterial culture than lichostatinal. Bromme explains that because the crystallography approach relies primarily on a protein’s affinity for an inhibitor, it automatically captures the most potent ones, even if they are present at lower concentrations than other less potent inhibitors.
“One question chemists always ask is, ‘What are we missing?’” says Timothy S. Bugni of the University of Wisconsin, Madison. With Bromme’s clever approach, he says, scientists can feel confident that they are pulling the most potent inhibitors out of nature. The approach won’t work on protein targets that aren’t readily crystallized, but for cases amenable to crystallography, Bugni says, it eliminates some of the most challenging steps—like determining a molecule’s chiral configuration during the structural characterization process—on the path from nature to pharmacy.
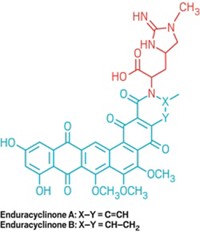
CORRECTION: This story was updated on Aug. 29, 2016, to correct the structure of lichostatinal.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter