Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Synthetic Biology
Artificial cofactor system sustains central metabolism in bacteria
System combines engineered glucose dehydrogenase and nicotinamide mononucleotide to achieve specificity
by Celia Henry Arnaud
December 3, 2019
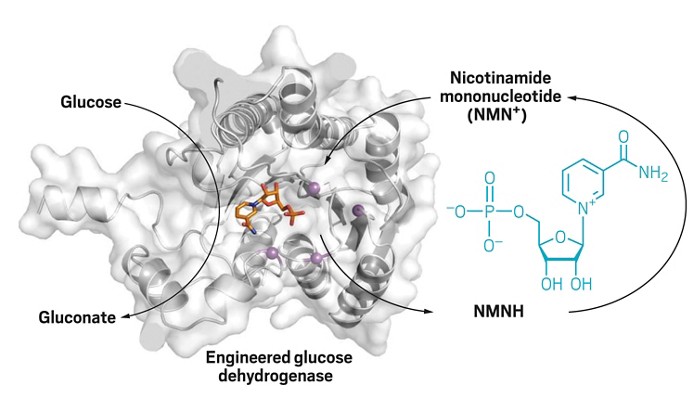
Many enzymes require cofactors such as nicotinamide adenosine dinucleotide (NAD+) or nicotinamide adenosine dinucleotide phosphate (NADP+) to shuttle electrons between reactions. Artificial redox cofactors that can work in parallel to the natural redox systems, without interfering with endogenous reactions, would enable improved metabolic engineering of industrially relevant compounds. Now, Han Li of the University of California Irvine, Justin B. Siegel of the University of California Davis, and coworkers have designed a new artificial redox cofactor system based on nicotinamide mononucleotide (NMN+) that is much more efficient than previous attempts (Nat. Chem. Biol. 2019, DOI: 10.1038/s41589-019-0402-7).
To make the system work, the researchers designed new versions of glucose dehydrogenase, an important partner for the cofactor. Their engineered version has more electrostatic interactions with NMN+ and thus recognizes NMN+ instead of the other cofactors. “The cofactor specificity is mainly engineered in the glucose dehydrogenase because that’s the enzyme that charges the cofactor with electrons,” Li says.
In one engineered version of glucose dehydrogenase, the researchers changed three amino acids. In the other variant, they changed four amino acids. The swapped amino acids improve the enzyme’s interactions with NMN+ and help it exclude NAD+ and NADP+.
NMN+ made a particularly good artificial cofactor because, compared with previous artificial cofactors, the molecule is large and has several “handles,” molecular features that enzymes can be engineered to grab onto in order to generate new interactions, Siegel says. Other artificial cofactors either have fewer molecular features for engineered enzymes to interact with, or they resemble the natural cofactors too much, making it difficult to exclude them. “This was really a diamond-in-the-rough cofactor,” he says.
As a proof of concept, the researchers coupled the cofactor and their engineered glucose dehydrogenase with an enoate reductase called XenA from Pseudomonas putida. This enzyme catalyzes the asymmetric reduction of the C=C double bond in ketoisopheorone to form levodione, a pharmaceutical intermediate. They also paired the engineered glucose dehydrogenase and NMN+ with three other enzymes, all of which worked.
The triple mutant glucose dehydrogenase works in test tubes but it interferes with other reactions in living systems because it doesn’t do a good enough job of excluding NAD+ and NADP+. The one with four mutations sufficiently excludes the natural cofactors to be used in living systems without disrupting the basic metabolism.
The researchers also showed that the four-mutation cofactor system can be used to sustain central metabolism in engineered bacteria. It’s the first artificial cofactor that’s efficient enough to do that, Li says. “Central carbon metabolism is like the highway of metabolism, so it’s got the highest flux. To sustain that, the system needs to be very efficient,” she says. “We show that you can actually use our system during glycolysis to support growth, which suggests it’s very efficient.”
Zongbao Kent Zhao of the Dalian Institute of Chemical Physics, who is also developing artificial cofactors, says the new research “opens new opportunities to devise orthogonal and efficient redox systems for biocatalysis and metabolic engineering.”



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter