Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Physical Chemistry
How To Salvage Rejected X-ray Crystallography Data
Including diffraction data that’s usually thrown away could improve crystal structures
by Carmen Drahl
May 28, 2012
| A version of this story appeared in
Volume 90, Issue 22
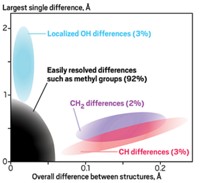
During their quests for biomolecule structures, X-ray crystallographers toss out data that’s useful after all, two independent teams contend in the journal Science. They report strategies for using traditionally ignored information to improve structures’ quality. When solving an unfamiliar protein’s structure, crystallographers obtain phase information to interpret X-ray diffraction patterns. For that task, they normally employ heavy atoms, such as selenium, but this isn’t always straightforward. For four different proteins, Qun Liu of the New York Structural Biology Center, Wayne A. Hendrickson of Columbia University, and colleagues combined data from multiple crystals, demonstrating that typically neglected information in a protein’s own sulfur atoms provides phase assistance with no heavy atoms needed (DOI: 10.1126/science.1218753). P. Andrew Karplus of Oregon State University and Kay Diederichs of the University of Konstanz, Germany, developed a statistical quantity called CC*. Compared with established criteria, CC* better tells researchers at what point the outer edges of diffraction patterns should be cut from calculations, the authors say (DOI: 10.1126/science.1218231). Pattern edges give the highest resolution information about atomic positions but can get clouded by noise, a tough trade-off that the authors suggest CC* resolves.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter