Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
ACS Meeting News
CRISPR is inefficient. Targeting a DNA repair protein could help.
Inhibiting DNA repair may improve CRISPR-Cas9 experiments
by Megha Satyanarayana
August 24, 2021
A small molecule could make the CRISPR-Cas9 gene editing tool more efficient, increasing the success of gene insertion experiments and streamlining lab work, according to work by medicinal chemist Lindsey James and graduate student Caroline Foley of the University of North Carolina at Chapel Hill.

Foley presented the work on Sunday in a Division of Medicinal Chemistry session at the American Chemical Society Fall 2021 meeting.
The CRISPR-Cas9 system is revolutionizing biology, but it’s inefficient. The system inserts DNA at the desired location less than 20% of the time. One reason CRISPR can fail is that when the Cas enzyme breaks DNA to insert genes of interest, the cell’s DNA repair mechanism fixes the break before the successful insertion is complete.
If CRISPR were even twice as efficient, James says, it could mean faster and more successful experiments.
Cells have a couple of mechanisms that help sew up DNA after the CRISPR-based insertion, but James says one of the mechanisms does a better job than the other. Blocking the other, somewhat sloppier mechanism could increase the probability of a successful DNA insertion.
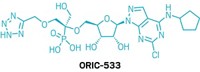
Their target is a protein called p53 binding protein 1, or 53BP1, which binds to methylated lysines on chromatin and facilitates the sloppier DNA repair mechanism. It’s been difficult to find a molecule that can block its action, James said, making it a challenging drug target. The team started with a small molecule called UNC 2170 but it had low affinity for the target protein, so in the new work, Foley screened 1,200 molecule fragments via a fluorescence-based screen to find a good starting block for a 53BP1 inhibitor. Her early results include a compound with an acyl piperazine moiety. Foley says they are continuing work to improve affinity of promising compounds.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter