Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Enzyme by Design
Computational design and directed evolution convert receptor protein into enzyme
by CELIA HENRY
July 12, 2004
| A version of this story appeared in
Volume 82, Issue 28

RETRACTION
The paper described in this article has since been retracted
(Science, DOI: 10.1126/science.319.5863.569b).
Computational design and directed evolution convert receptor protein into enzyme
The true test of understanding a reaction is creating an enzyme from scratch to catalyze it. Biochemists at Duke University Medical Center now have shown that they can turn a protein having no catalytic activity into an enzyme that speeds up a reaction that is unrelated to the protein's original function.
Through a combination of computational design and directed evolution, biochemistry professor Homme W. Hellinga, working with graduate students Mary A. Dwyer and Loren L. Looger, turned the receptor ribose-binding protein (RBP) into an enzyme that mimics the natural enzyme triose phosphate isomerase (TIM) [Science, 304, 1967 (2004)].
"This is one of the very first times that we have been able to design, essentially from first principles, an enzymatic reaction using structure-based design," Hellinga says. "You can dial in a particular reaction mechanism and turn a protein that normally doesn't do anything into a little enzyme."
The team chose their example carefully, selecting a well-understood reaction. TIM catalyzes the interconversion of dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP) through an isomerization reaction. TIM participates in glycolysis (the breakdown of glucose) and in gluconeogenesis (the production of glucose from noncarbohydrate sources).
In a starting protein, the so-called scaffold protein must be able to accommodate the reaction in terms of molecular volume and geometry. "The only thing you could say is a great similarity [between RBP and TIM] is that ribose-binding protein has a cavity that would normally bind ribose that's large enough to hold the substrate of triose phosphate isomerase," he says. "That's the only sort of decision that you make when you choose a particular scaffold."
Some protein-to-enzyme transformations simply won't be possible. For example, Hellinga notes, "there's no way you're going to turn ribose-binding protein into a DNA polymerase."
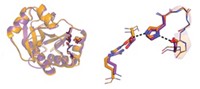
The two main challenges in designing the enzyme were describing the problem and solving the resulting combinatorial search problem. The "description problem" involves "capturing the essential elements of the reaction mechanism," Hellinga says. Such descriptions include the type of reaction, the transition state, the geometry of the reaction, and ways to stabilize the transition state.
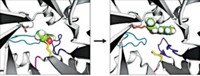
The Duke researchers converted that description into algorithms that introduce mutations in the three-dimensional protein structure. They needed to specify the amino acid residues involved in the catalysis and to identify mutations that would facilitate interaction with the substrate and stabilize the protein.
"When you do these kinds of calculations, you're faced with many different choices," Hellinga says. "In the end, we had 21 or 22 mutations that we played with, which is a huge computational search problem."
The researchers started by testing whether they could redesign RBP to bind TIM's substrates, regardless of catalytic activity. They changed the layer of amino acids that directly contact ribose in the normal protein and found four designs that bind both DHAP and GAP. The proteins were constructed by mutagenesis in bacteria.
To introduce catalytic activity, they carefully placed amino acids with reactive side chains, such as glutamate, histidine, and lysine, within the active site. Seven of 14 designs showed increases in GAP production compared with background levels, and one design, which they dubbed NovoTim1.0, was significantly more active. NovoTim1.0, which was thermally unstable, was then used as a starting point to design proteins that were more stable.
SOME DIFFICULTIES with the reaction required Hellinga to improve the enzymes using directed evolution. The most common carbon sources for gluconeogenesis are lactate and glycerol; glycerol requires more enzyme activity.
"Our designed proteins grew on lactate but not on glycerol," Hellinga says. "We did a directed evolution experiment using our selection method to ask for variants that were obtained randomly and that would grow on glycerol. It turned out that a tiny tweak in activity was needed."
In an accompanying commentary [Science, 304, 1916 (2004)], German researchers Reinhard Sterner from the Institute for Biophysics & Physical Biochemistry at the University of Regensburg and Franz X. Schmid of the Laboratory for Biochemistry at the University of Bayreuth write: "The new work by the Hellinga lab exemplifies the enormous power of computational biology and illustrates how this approach can be combined with directed evolution. The latter is well suited to identify beneficial mutations far from the active site. Such mutations are difficult to find by computation but important for the fine-tuning of catalysis."
Hellinga hopes to be able eventually to design any enzyme at will. "I'm sure everybody's reaction to that is going to be 'Good luck,' " he says. First, he needs to find out if his group can do this again with another reaction or if the first success was just a fluke. His group members have picked other reactions and have started to see if they can build enzymes for them.
"We're quietly optimistic that this is a very nice way to go," Hellinga says. "My guess is that computational design in the near term is not going to get you all the way, meaning you'll get pretty good enzymes but not very good enzymes. If you can then marry that with clever directed evolution methods, it's like a marriage made in heaven. It will go a long, long way."


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter