Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Protein Sequencing for the Masses
High-end MS sequencing method is adapted for more affordable benchtop mass spectrometers
by STU BORMAN, C&EN WASHINGTON
July 12, 2004
| A version of this story appeared in
Volume 82, Issue 28

Mass Spectrometry (MS) is of key importance in proteomics, the study of protein structure and function on a genome-wide scale. In proteomics studies, one typically wants to identify a protein by using MS to sequence peptide fragments from the protein. But such analyses often fail for proteins whose peptides contain phosphates and other modifications, multiple arginine residues, or more than 20 to 25 amino acids. The only way around this problem has been to use a technique called electron capture dissociation (ECD) on an expensive type of mass spectrometer that is not widely available in proteomics labs.
Now, chemistry and pathology professor Donald F. Hunt of the University of Virginia and coworkers at the university and at Thermo Electron, San Jose, Calif., have adapted ECD for use on one of the least expensive and most widely available types of mass spectrometer [Proc. Natl. Acad. Sci. USA, 101, 9528 (2004)]. The technique they developed is called electron transfer dissociation (ETD). "It is my belief that ETD will revolutionize how mass spectrometry is applied to the field of proteomics," Hunt says.
That might seem self-serving, but chemistry professor Scott A. McLuckey of Purdue University, who specializes in peptide and protein sequencing by MS, basically agrees. "We'll have to see how it plays out, but I think ETD is going to have a big impact," he says. "It's going to be very useful because it opens up this kind of sequence information to a much larger community. For me, it's the most interesting development of the last couple of years in proteomics."
Professor of biochemistry and molecular biology Matthias Mann of the University of Southern Denmark, who specializes in MS-based proteomics, says: "This is very nice and potentially revolutionary work. Particularly for modified peptides, it could indeed be a major step forward. All the examples in the PNAS paper are triply charged, and in proteomics experiments the majority of peptides are usually less than triply charged. So I think we need to see a little more data, but if that works out we will be clamoring to have this technology on our instrument, too."
ETD is an extension of ECD, which was developed by chemistry professor emeritus Fred W. McLafferty of Cornell University and coworkers [J. Am. Chem. Soc., 120, 3265 (1998)]. In ECD, electrons captured by multiply charged peptide or protein cations cause the structures to fragment nonselectively (in a sequence independent manner) at peptide bonds, making it possible to obtain extensive sequence information. But ECD could only be carried out on Fourier transform ion cyclotron resonance (FT-ICR) spectrometers, which have superconducting magnets of high magnetic field strength and are the most expensive type of MS instruments.
"Most people who do proteomics don't have such instruments," McLuckey says, "so the ECD technique has not been particularly widely accessible."
Ion trap mass spectrometers are available as inexpensive benchtop units and are therefore found in many more labs. Peptides and proteins also can be analyzed in ion trap spectrometers using collisionally activated dissociation, in which the molecules are fragmented by collisions with an inert gas, such as helium or argon atoms. Such fragmentation is sequence dependent, however, with the weakest bonds being cleaved, so that in many cases one doesn't obtain sufficient useful structural information from this type of experiment alone. In any case, the McLafferty group's ECD technique could not be carried out in ion trap spectrometers because the thermal electrons needed for the electron capture process don't retain their thermal energy for a long enough time in such instruments.
HUNT AND COWORKERS have now devised the ETD process, in which electrons are instead transferred from singly charged anthracene anions to positively charged peptides or proteins. This circumvents the thermal electron problem and makes it possible to use ECD (with the ETD modification) in widely available ion trap instruments. "We had been looking for such a reagent and hadn't found one," McLuckey says.
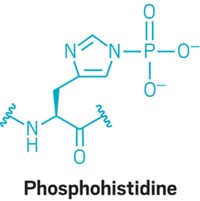
Many natural proteins are phosphorylated, and researchers would like to know the phosphorylation sites. With conventional MS fragmentation methods, such as collisionally activated dissociation, the phosphate groups fall off, and the sites where they were located can't be determined. ETD, on the other hand, cleaves the polyamide backbone of peptide and protein cations, instead of excising side chains such as phosphate groups, making it possible to identify phosphorylation sites and making ETD applicable to the analysis of phosphorylated proteins.
PEPTIDE AND PROTEIN phosphorylation mapping can also be carried out by liquid chromatography-tandem MS or by a combined chemical-enzymatic approach devised by professor of cellular and molecular pharmacology Kevan M. Shokat of the University of California, San Francisco, and coworkers [Nat. Biotechnol., 21, 1047 (2003); C&EN, Sept. 1, 2003, page 31]. But such specialized techniques require extra steps apart from the usual sequencing experiments, so ETD may prove more generally useful for identifying phosphorylated peptides and proteins.
"If ETD extrapolates the way one would hope," McLuckey says, "it also might be a very nice way to characterize other posttranslational modifications" in addition to phosphates, such as carbohydrate groups in glycosylated proteins, "because usually when you fragment posttranslationally modified proteins they lose the modifications," whereas with ETD they do not. Hunt and coworkers also believe that it may be possible to carry out "top-down" analyses--direct sequencing of proteins, without prior enzymatic fragmentation--by combining ETD with collisionally activated dissociation and ion-ion proton-transfer techniques developed earlier by McLuckey's group.
Overall, Hunt and coworkers conclude that ETD will become "an indispensable tool for peptide and protein sequence analysis in the near future and is likely to drive the development of new MS instrumentation and software."
McLafferty agrees. "Having the same capabilities as ECD on a far less expensive instrument already in constant use in proteomics laboratories will be a great boon," he says.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter