Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Repair Enzyme Flips and Snips
Structure fills out picture of how cells carry out error-free repair of DNA
by Amanda Yarnell
February 16, 2004
| A version of this story appeared in
Volume 82, Issue 7

The process by which cells correct the damage done to DNA by reactive oxygen species just got a little clearer, thanks to a new structure of the repair enzyme MutY bound to its damaged substrate.
Produced by everything from smoking to normal metabolic activity, reactive oxygen species are constantly damaging the nucleotide bases that make up genomic DNA. One of the most common types of damage is the conversion of guanine to 8-oxoguanine (8-oxoG). If allowed to persist, 8-oxoG lesions will pair with adenine--not cytosine, guanine's normal partner--during DNA replication.
8-OxoG:A mispairs eventually give rise to dangerous genetic mutations, so cells have devised a way to repair both bases to restore the original DNA sequence. MutY carries out the first step of this process: finding 8-oxoG:A mispairs and removing the adenine.
"How MutY distinguishes A residues paired abnormally with 8-oxoG amidst the vast excess of A residues paired normally with T [thymine] has been a long-standing question in the field," notes Harvard University chemistry professor Gregory L. Verdine.
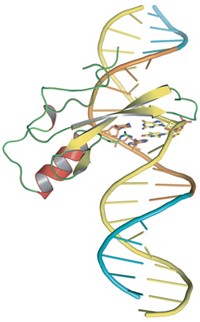
Verdine, graduate student J. Christopher Fromme, and their coworkers have now neatly answered this question by solving a 2.2-Å X-ray structure of MutY bound to DNA containing an 8-oxoG:A mispair [Nature, 427, 652 (2004)]. None of the half dozen hydrogen bonds that the enzyme makes to the 8-oxoG in this structure would be possible with a thymine, Verdine points out.
The structure also reveals how MutY removes the unwanted adenine. MutY creates a sharp bend in the DNA at the site of the 8-oxoG:A mispair. It then extrudes the adenine into its active site and clips the base from the DNA backbone.
Calling the research a "tour de force," chemistry professor Sheila S. David of the University of Utah notes that Verdine's team used a number of clever tricks to stabilize the normally fragile complex, including using a hardy version of MutY from a thermophilic bacterium and cross-linking the enzyme to the DNA substrate via a disulfide bond.
A team led by David previously showed that two different inherited mutations in Myh--the human version of MutY--predispose patients to colon cancer. She points out that the new structure explains why these mutations impair Myh's activity, allowing cancer-causing 8-oxoG:A mispairs to persist. In both cases, the amino acid that's mutated normally plays an important role in recognizing 8-oxoG, Verdine's team reports.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter